6WKT
 
 | |
3GZT
 
 | | VP7 recoated rotavirus DLP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Outer capsid glycoprotein VP7 | | Authors: | Chen, J.Z, Settembre, E.C, Harrison, S.C, Grigorieff, N. | | Deposit date: | 2009-04-07 | | Release date: | 2009-07-14 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular interactions in rotavirus assembly and uncoating seen by high-resolution cryo-EM
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GZU
 
 | | VP7 recoated rotavirus DLP | | Descriptor: | Inner capsid protein VP2, Intermediate capsid protein VP6, ZINC ION | | Authors: | Chen, J.Z, Settembre, E.C, Harrison, S.C, Grigorieff, N. | | Deposit date: | 2009-04-07 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular interactions in rotavirus assembly and uncoating seen by high-resolution cryo-EM.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4V7Q
 
 | | Atomic model of an infectious rotavirus particle | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Core scaffold protein, ... | | Authors: | Settembre, E.C, Chen, J.Z, Dormitzer, P.R, Grigorieff, N, Harrison, S.C. | | Deposit date: | 2010-05-13 | | Release date: | 2014-07-09 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic model of an infectious rotavirus particle.
Embo J., 30, 2011
|
|
4AU6
 
 | | Location of the dsRNA-dependent polymerase, VP1, in rotavirus particles | | Descriptor: | RNA-DEPENDENT RNA POLYMERASE | | Authors: | Estrozi, L.F, Settembre, E.C, Goret, G, McClain, B, Zhang, X, Chen, J.Z, Grigorieff, N, Harrison, S.C. | | Deposit date: | 2012-05-14 | | Release date: | 2012-06-13 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Location of the Dsrna-Dependent Polymerase, Vp1, in Rotavirus Particles.
J.Mol.Biol., 425, 2013
|
|
4F5X
 
 | | Location of the dsRNA-dependent polymerase, VP1, in rotavirus particles | | Descriptor: | Intermediate capsid protein VP6, RNA-directed RNA polymerase, VP2 protein, ... | | Authors: | Estrozi, L.F, Settembre, E.C, Goret, G, McClain, B, Zhang, X, Chen, J.Z, Grigorieff, N, Harrison, S.C. | | Deposit date: | 2012-05-13 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Location of the dsRNA-Dependent Polymerase, VP1, in Rotavirus Particles.
J.Mol.Biol., 425, 2013
|
|
2BGZ
 
 | | ATOMIC MODEL OF THE BACTERIAL FLAGELLAR BASED ON DOCKING AN X-RAY DERIVED HOOK STRUCTURE INTO AN EM MAP. | | Descriptor: | FLAGELLAR HOOK PROTEIN FLGE | | Authors: | Shaikh, T.R, Thomas, D.R, Chen, J.Z, Samatey, F.A, Matsunami, H, Imada, K, Namba, K, Derosier, D.J. | | Deposit date: | 2005-01-06 | | Release date: | 2005-01-27 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | A Partial Atomic Structure for the Flagellar Hook of Salmonella Typhimurium.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BGY
 
 | | Fit of the x-ray structure of the baterial flagellar hook fragment flge31 into an EM map from the hook of Caulobacter crescentus. | | Descriptor: | FLAGELLAR HOOK PROTEIN FLGE | | Authors: | Shaikh, T.R, Thomas, D.R, Chen, J.Z, Samatey, F.A, Matsunami, H, Imada, K, Namba, K, DeRosier, D.J. | | Deposit date: | 2005-01-06 | | Release date: | 2005-01-27 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | A Partial Atomic Structure for the Flagellar Hook of Salmonella Typhimurium.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
5TWV
 
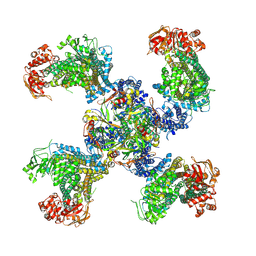 | | Cryo-EM structure of the pancreatic ATP-sensitive K+ channel SUR1/Kir6.2 in the presence of ATP and glibenclamide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ATP-sensitive inward rectifier potassium channel 11 | | Authors: | Martin, G.M, Yoshioka, C, Chen, J.Z, Shyng, S.L. | | Deposit date: | 2016-11-14 | | Release date: | 2017-01-25 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Cryo-EM structure of the ATP-sensitive potassium channel illuminates mechanisms of assembly and gating.
Elife, 6, 2017
|
|
6MYX
 
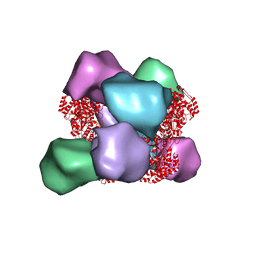 | | EM structure of Bacillus subtilis ribonucleotide reductase inhibited double-helical filament of NrdE alpha subunit with dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Ribonucleoside-diphosphate reductase | | Authors: | Thomas, W.C, Bacik, J.P, Chen, J.Z, Ando, N. | | Deposit date: | 2018-11-02 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
4TVS
 
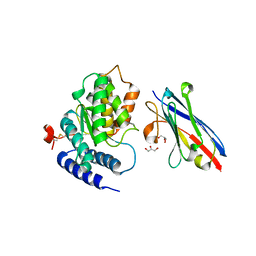 | | LAP1(aa356-583), H.sapiens, bound to VHH-BS1 | | Descriptor: | GLYCEROL, SULFATE ION, Torsin-1A-interacting protein 1, ... | | Authors: | Sosa, B.A, Schwartz, T.U. | | Deposit date: | 2014-06-27 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | How lamina-associated polypeptide 1 (LAP1) activates Torsin.
Elife, 3, 2014
|
|
6MW3
 
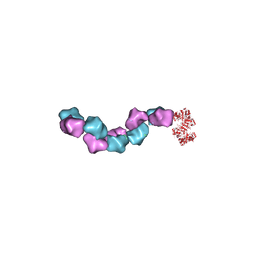 | | EM structure of Bacillus subtilis ribonucleotide reductase inhibited filament composed of NrdE alpha subunit and NrdF beta subunit with dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Ribonucleoside-diphosphate reductase, Ribonucleoside-diphosphate reductase NrdF beta subunit | | Authors: | Thomas, W.C, Bacik, J.P, Kaelber, J.T, Ando, N. | | Deposit date: | 2018-10-29 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.65 Å) | | Cite: | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
1WLG
 
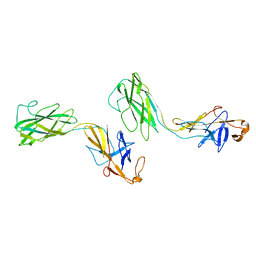 | | Crystal structure of FlgE31, a major fragment of the hook protein | | Descriptor: | Flagellar hook protein flgE | | Authors: | Samatey, F.A, Matsunami, H, Imada, K, Nagashima, S, Shaikh, T.R, Thomas, D.R, DeRosier, D.J, Kitao, A, Namba, K. | | Deposit date: | 2004-06-25 | | Release date: | 2004-11-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the bacterial flagellar hook and implication for the molecular universal joint mechanism.
Nature, 431, 2004
|
|
6D3S
 
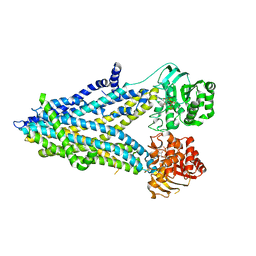 | | Thermostabilized phosphorylated chicken CFTR | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator | | Authors: | Fay, J.F, Riordan, J.R, Chen, Z.J. | | Deposit date: | 2018-04-16 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Cryo-EM Visualization of an Active High Open Probability CFTR Anion Channel.
Biochemistry, 57, 2018
|
|
2OM3
 
 | | High-resolution cryo-EM structure of Tobacco Mosaic Virus | | Descriptor: | Coat protein, Tobacco Mosaic Virus RNA | | Authors: | Sachse, C. | | Deposit date: | 2007-01-20 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | High-resolution electron microscopy of helical specimens: a fresh look at tobacco mosaic virus.
J.Mol.Biol., 371, 2007
|
|
6D3R
 
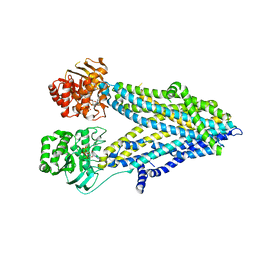 | | Thermostablilized dephosphorylated chicken CFTR | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator | | Authors: | Fay, J.F, Riordan, J.R. | | Deposit date: | 2018-04-16 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM Visualization of an Active High Open Probability CFTR Anion Channel.
Biochemistry, 57, 2018
|
|
4JIA
 
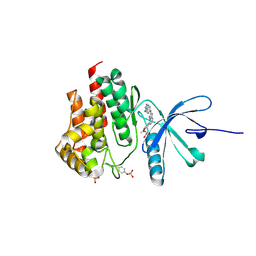 | | JAK2 kinase (JH1 domain) in complex with compound 9 | | Descriptor: | 5-(4-methoxyphenyl)-N-[4-(4-methylpiperazin-1-yl)phenyl][1,2,4]triazolo[1,5-a]pyridin-2-amine, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2013-03-05 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 2-Amino-[1,2,4]triazolo[1,5-a]pyridines as JAK2 inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4JI9
 
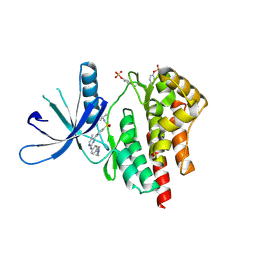 | | JAK2 kinase (JH1 domain) in complex with TG101209 | | Descriptor: | N-tert-butyl-3-[(5-methyl-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino]benzenesulfonamide, Tyrosine-protein kinase JAK2 | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2013-03-05 | | Release date: | 2013-08-07 | | Last modified: | 2013-08-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2-Amino-[1,2,4]triazolo[1,5-a]pyridines as JAK2 inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4KF7
 
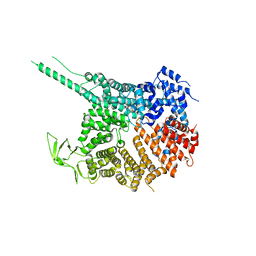 | |
4KF8
 
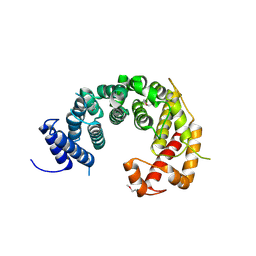 | |
6MT9
 
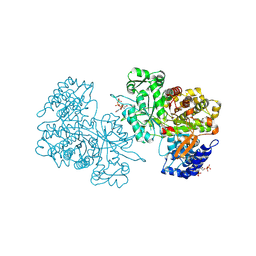 | | X-ray crystal structure of Bacillus subtilis ribonucleotide reductase NrdE alpha subunit with TTP, ATP, and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Thomas, W.C, Brooks, F.P, Bacik, J.P, Ando, N. | | Deposit date: | 2018-10-19 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
6MV9
 
 | | X-ray crystal structure of Bacillus subtilis ribonucleotide reductase NrdE alpha subunit with TTP and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ribonucleoside-diphosphate reductase, ... | | Authors: | Thomas, W.C, Brooks, F.P, Bacik, J.P, Ando, N. | | Deposit date: | 2018-10-24 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
6MVE
 
 | | Reduced X-ray crystal structure of Bacillus subtilis ribonucleotide reductase NrdE alpha subunit with TTP, ATP, and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Thomas, W.C, Brooks, F.P, Bacik, J.P, Ando, N. | | Deposit date: | 2018-10-25 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
