4GU0
 
 | | Crystal structure of LSD2 with H3 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Histone H3.3, Lysine-specific histone demethylase 1B, ... | | Authors: | Chen, F, Yang, H, Dong, Z, Fang, J, Zhu, T, Gong, W, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Structural insight into substrate recognition by histone demethylase LSD2/KDM1b
Cell Res., 23, 2013
|
|
4HSU
 
 | | Crystal structure of LSD2-NPAC with H3(1-26)in space group P21 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Histone H3, Lysine-specific histone demethylase 1B, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Xu, Y. | | Deposit date: | 2012-10-30 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.988 Å) | | Cite: | Structural insight into substrate recognition by histone demethylase LSD2/KDM1b.
Cell Res., 23, 2013
|
|
6R6U
 
 | | Crystal structure of human cis-aconitate decarboxylase | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Cis-aconitate decarboxylase, SODIUM ION | | Authors: | Lukat, P, Chen, F, Saile, K, Buessow, K, Pessler, F, Blankenfeldt, W. | | Deposit date: | 2019-03-28 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | Crystal structure ofcis-aconitate decarboxylase reveals the impact of naturally occurring human mutations on itaconate synthesis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6R6T
 
 | | Crystal structure of mouse cis-aconitate decarboxylase | | Descriptor: | Cis-aconitate decarboxylase | | Authors: | Lukat, P, Chen, F, Saile, K, Buessow, K, Pessler, F, Blankenfeldt, W. | | Deposit date: | 2019-03-28 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.535 Å) | | Cite: | Crystal structure ofcis-aconitate decarboxylase reveals the impact of naturally occurring human mutations on itaconate synthesis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3VAX
 
 | | Crystal structure of DndA from streptomyces lividans | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative uncharacterized protein dndA | | Authors: | Zhang, Z, Chen, F, Lin, K, Wu, G. | | Deposit date: | 2011-12-30 | | Release date: | 2013-01-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the cysteine desulfurase DndA from Streptomyces lividans which is involved in DNA phosphorothioation
Plos One, 7, 2012
|
|
4GUS
 
 | | Crystal structure of LSD2-NPAC with H3 in space group P3221 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Histone H3.3, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4GUR
 
 | | Crystal structure of LSD2-NPAC with H3 in space group P21 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Histone H3.3, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4GUT
 
 | | Crystal structure of LSD2-NPAC | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1B, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4GU1
 
 | | Crystal structure of LSD2 | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1B, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.939 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4GUU
 
 | | Crystal structure of LSD2-NPAC with tranylcypromine | | Descriptor: | Lysine-specific histone demethylase 1B, Putative oxidoreductase GLYR1, ZINC ION, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
3KYS
 
 | | Crystal structure of human YAP and TEAD complex | | Descriptor: | 65 kDa Yes-associated protein, Transcriptional enhancer factor TEF-1 | | Authors: | Li, Z, Zhao, B, Wang, P, Chen, F, Dong, Z, Yang, H, Guan, K.L, Xu, Y. | | Deposit date: | 2009-12-07 | | Release date: | 2010-02-23 | | Last modified: | 2020-10-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the YAP and TEAD complex
Genes Dev., 24, 2010
|
|
2A7R
 
 | | Crystal structure of human Guanosine Monophosphate reductase 2 (GMPR2) | | Descriptor: | GMP reductase 2, GUANOSINE-5'-MONOPHOSPHATE, SULFATE ION | | Authors: | Li, J, Wei, Z, Zheng, M, Gu, X, Deng, Y, Qiu, R, Chen, F, Ji, C, Gong, W, Xie, Y, Mao, Y. | | Deposit date: | 2005-07-05 | | Release date: | 2006-01-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Human Guanosine Monophosphate Reductase 2 (GMPR2) in Complex with GMP
J.Mol.Biol., 355, 2006
|
|
4GY5
 
 | | Crystal structure of the tandem tudor domain and plant homeodomain of UHRF1 with Histone H3K9me3 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Peptide from Histone H3.3, ZINC ION | | Authors: | Cheng, J, Yang, Y, Fang, J, Xiao, J, Zhu, T, Chen, F, Wang, P, Xu, Y. | | Deposit date: | 2012-09-05 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.956 Å) | | Cite: | Structural insight into coordinated recognition of trimethylated histone H3 lysine 9 (H3K9me3) by the plant homeodomain (PHD) and tandem tudor domain (TTD) of UHRF1 (ubiquitin-like, containing PHD and RING finger domains, 1) protein
J.Biol.Chem., 288, 2013
|
|
7VRB
 
 | | Structure of the Human BRG1/SS18 complex | | Descriptor: | SMARCA4 protein,Protein SSXT | | Authors: | Cheng, Y, Chen, F, Zhou, H, Long, J. | | Deposit date: | 2021-10-22 | | Release date: | 2022-05-25 | | Last modified: | 2022-12-07 | | Method: | X-RAY DIFFRACTION (2.389 Å) | | Cite: | Phase transition and remodeling complex assembly are important for SS18-SSX oncogenic activity in synovial sarcomas.
Nat Commun, 13, 2022
|
|
7VRC
 
 | |
4RQU
 
 | | Alcohol Dehydrogenase crystal structure in complex with NAD | | Descriptor: | Alcohol Dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Xu, Y.W. | | Deposit date: | 2014-11-05 | | Release date: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insight into the conformational change of alcohol dehydrogenase from arabidopsis Thalianal during coenzyme binding.
Biochimie, 108C, 2014
|
|
4RQT
 
 | | Alcohol Dehydrogenase Crystal Structure | | Descriptor: | ACETIC ACID, Alcohol dehydrogenase class-P, SULFATE ION, ... | | Authors: | Xu, Y.W. | | Deposit date: | 2014-11-05 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into the conformational change of alcohol dehydrogenase from arabidopsis Thalianal during coenzyme binding.
Biochimie, 108C, 2014
|
|
1VKX
 
 | | CRYSTAL STRUCTURE OF THE NFKB P50/P65 HETERODIMER COMPLEXED TO THE IMMUNOGLOBULIN KB DNA | | Descriptor: | DNA (5'-D(*AP*GP*GP*AP*AP*AP*GP*TP*CP*CP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*GP*GP*AP*CP*TP*TP*TP*CP*C)-3'), PROTEIN (NF-KAPPA B P50 SUBUNIT), ... | | Authors: | Chen, F, Huang, D.B, Ghosh, G. | | Deposit date: | 1997-09-17 | | Release date: | 1998-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA.
Nature, 391, 1998
|
|
8H41
 
 | | Crystal structure of a decarboxylase from Trichosporon moniliiforme in complex with o-nitrophenol | | Descriptor: | MAGNESIUM ION, O-NITROPHENOL, Salicylate decarboxylase | | Authors: | Gao, J, Zhao, Y.P, Li, Q, Liu, W.D, Sheng, X. | | Deposit date: | 2022-10-09 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A Combined Computational-Experimental Study on the Substrate Binding and Reaction Mechanism of Salicylic Acid Decarboxylase
Catalysts, 12, 2022
|
|
3QCW
 
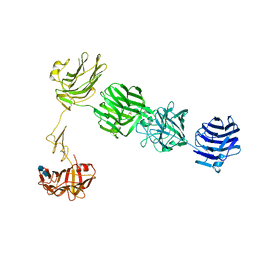 | | Structure of neurexin 1 alpha (domains LNS1-LNS6), no splice inserts | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Neurexin-1-alpha | | Authors: | Rudenko, G. | | Deposit date: | 2011-01-17 | | Release date: | 2011-06-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Structure of Neurexin 1 alpha Reveals Features Promoting a Role as Synaptic Organizer
Structure, 19, 2011
|
|
3R05
 
 | |
5TSL
 
 | |
5TSK
 
 | |
6U6H
 
 | | Calcium-bound MthK open-inactivated state 3 | | Descriptor: | Calcium-gated potassium channel MthK | | Authors: | Chen, F, Crina, N. | | Deposit date: | 2019-08-29 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Ball-and-chain inactivation in a calcium-gated potassium channel.
Nature, 580, 2020
|
|
6UXB
 
 | |
