4WIZ
 
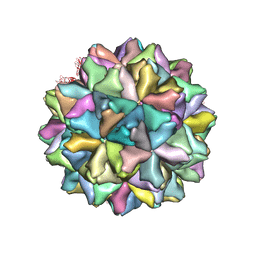 | | Crystal structure of Grouper nervous necrosis virus-like particle at 3.6A | | Descriptor: | CALCIUM ION, Coat protein | | Authors: | Chen, N.C, Chen, C.J, Yoshimura, M, Guan, H.H, Chen, T.Y. | | Deposit date: | 2014-09-28 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal Structures of a Piscine Betanodavirus: Mechanisms of Capsid Assembly and Viral Infection
Plos Pathog., 11, 2015
|
|
4RFU
 
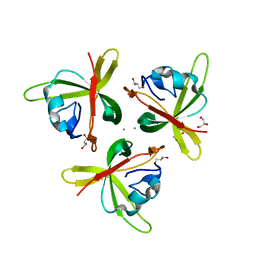 | | Crystal structure of truncated P-domain from Grouper nervous necrosis virus capsid protein at 1.2A | | Descriptor: | CALCIUM ION, Coat protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chen, N.C, Chen, C.J, Yoshimura, M, Guan, H.H, Chen, T.Y. | | Deposit date: | 2014-09-27 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structures of a Piscine Betanodavirus: Mechanisms of Capsid Assembly and Viral Infection
Plos Pathog., 11, 2015
|
|
4RFT
 
 | | T=1 subviral particle of Grouper nervous necrosis virus capsid protein deletion mutant (delta 1-34 & 218-338) | | Descriptor: | Coat protein | | Authors: | Chen, N.C, Chen, C.J, Yoshimura, M, Guan, H.H, Chen, T.Y. | | Deposit date: | 2014-09-27 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structures of a Piscine Betanodavirus: Mechanisms of Capsid Assembly and Viral Infection
Plos Pathog., 11, 2015
|
|
4YM4
 
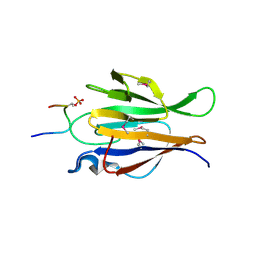 | | Truncated Human TIFA in complex with its Thr9 phosphorylated N-terminal peptide 1-15 | | Descriptor: | TRAF-interacting protein with FHA domain-containing protein A | | Authors: | Weng, J.H, Wei, T.Y.W, Hsieh, Y.C, Huang, C.C.F, Wu, P.Y.G, Chen, E.S.W, Huang, K.F, Chen, C.J, Tsai, M.D. | | Deposit date: | 2015-03-06 | | Release date: | 2015-10-21 | | Last modified: | 2015-10-28 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Uncovering the Mechanism of Forkhead-Associated Domain-Mediated TIFA Oligomerization That Plays a Central Role in Immune Responses.
Biochemistry, 54, 2015
|
|
4ZGI
 
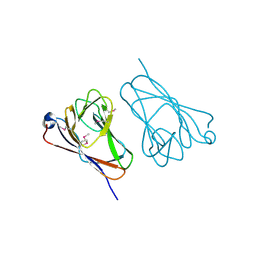 | | Structure of Truncated Human TIFA | | Descriptor: | TRAF-interacting protein with FHA domain-containing protein A | | Authors: | Weng, J.H, Wei, T.Y.W, Hsieh, Y.C, Huang, C.C.F, Wu, P.Y.G, Chen, E.S.W, Huang, K.F, Chen, C.J, Tsai, M.D. | | Deposit date: | 2015-04-23 | | Release date: | 2015-10-14 | | Last modified: | 2015-10-28 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Uncovering the Mechanism of Forkhead-Associated Domain-Mediated TIFA Oligomerization That Plays a Central Role in Immune Responses.
Biochemistry, 54, 2015
|
|
4XPX
 
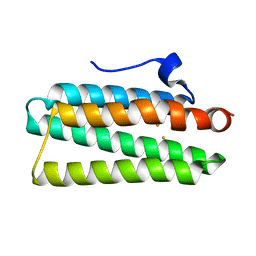 | | Crystal structure of hemerythrin:wild-type | | Descriptor: | Bacteriohemerythrin, FE (II) ION | | Authors: | Chuankhayan, P, Chen, K.H.C, Wu, H.H, Chen, C.J, Fukuda, M, Yu, S.S.F, Chan, S.I. | | Deposit date: | 2015-01-18 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | The bacteriohemerythrin from Methylococcus capsulatus (Bath): Crystal structures reveal that Leu114 regulates a water tunnel.
J.Inorg.Biochem., 150, 2015
|
|
4XQ1
 
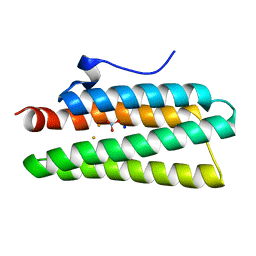 | | Crystal structure of hemerythrin: L114A mutant | | Descriptor: | Bacteriohemerythrin, FE (III) ION, NITRATE ION, ... | | Authors: | Chuankhayan, P, Chen, K.H.C, Wu, H.H, Chen, C.J, Fukuda, M, Yu, S.S.F, Chan, S.I. | | Deposit date: | 2015-01-18 | | Release date: | 2015-04-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The bacteriohemerythrin from Methylococcus capsulatus (Bath): Crystal structures reveal that Leu114 regulates a water tunnel.
J.Inorg.Biochem., 150, 2015
|
|
4XPW
 
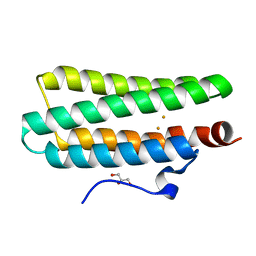 | | Crystal structures of Leu114F mutant | | Descriptor: | Bacteriohemerythrin, FE (II) ION, GLYCEROL | | Authors: | Chuankhayan, P, Chen, K.H.C, Wu, H.H, Chen, C.J, Fukuda, M, Yu, S.S.F, Chan, S.I. | | Deposit date: | 2015-01-18 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | The bacteriohemerythrin from Methylococcus capsulatus (Bath): Crystal structures reveal that Leu114 regulates a water tunnel.
J.Inorg.Biochem., 150, 2015
|
|
4XPY
 
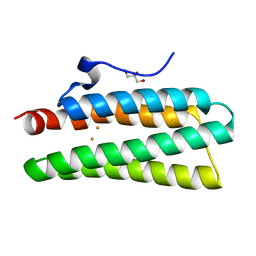 | | Crystal structure of hemerythrin : L114Y mutant | | Descriptor: | Bacteriohemerythrin, FE (II) ION, GLYCEROL | | Authors: | Chuankhayan, P, Chen, K.H.C, Wu, H.H, Chen, C.J, Fukuda, M, Yu, S.S.F, Chan, S.I. | | Deposit date: | 2015-01-18 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | The bacteriohemerythrin from Methylococcus capsulatus (Bath): Crystal structures reveal that Leu114 regulates a water tunnel.
J.Inorg.Biochem., 150, 2015
|
|
1ZOI
 
 | | Crystal Structure of a Stereoselective Esterase from Pseudomonas putida IFO12996 | | Descriptor: | esterase | | Authors: | Elmi, F, Lee, H.T, Huang, J.Y, Hsieh, Y.C, Wang, Y.L, Chen, Y.J, Shaw, S.Y, Chen, C.J. | | Deposit date: | 2005-05-13 | | Release date: | 2006-05-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Stereoselective esterase from Pseudomonas putida IFO12996 reveals alpha/beta hydrolase folds for D-beta-acetylthioisobutyric acid synthesis
J.Bacteriol., 187, 2005
|
|
2GJ5
 
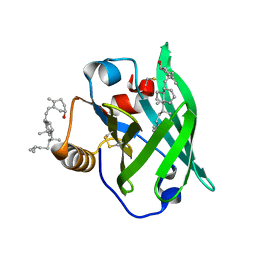 | | Crystal structure of a secondary vitamin D3 binding site of milk beta-lactoglobulin | | Descriptor: | (1S,3Z)-3-[(2E)-2-[(1R,3AR,7AS)-7A-METHYL-1-[(2R)-6-METHYLHEPTAN-2-YL]-2,3,3A,5,6,7-HEXAHYDRO-1H-INDEN-4-YLIDENE]ETHYLI DENE]-4-METHYLIDENE-CYCLOHEXAN-1-OL, Beta-lactoglobulin | | Authors: | Yang, M.C, Guan, H.H, Liu, M.Y, Yang, J.M, Chen, W.L, Chen, C.J, Mao, S.J. | | Deposit date: | 2006-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a secondary vitamin D3 binding site of milk beta-lactoglobulin.
Proteins, 71, 2008
|
|
3LFI
 
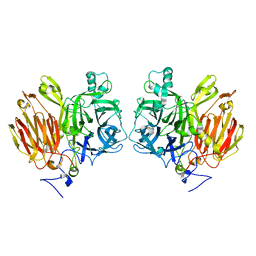 | |
3LDK
 
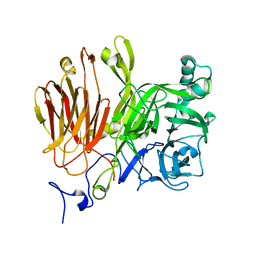 | | Crystal Structure of A. japonicus CB05 | | Descriptor: | Fructosyltransferase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Chuankhayan, P, Chen, C.J, Chaing, C.M, Hsieh, C.Y, Chen, C.D, Hsieh, Y.C. | | Deposit date: | 2010-01-13 | | Release date: | 2010-05-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Aspergillus japonicus fructosyltransferase complex with donor/acceptor substrates reveal complete sbusites in the active site for catalysis
To be Published
|
|
2QN4
 
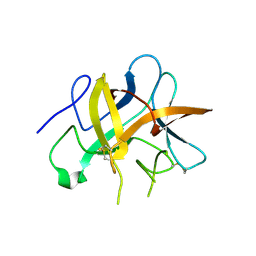 | | Structure and function study of rice bifunctional alpha-amylase/subtilisin inhibitor from Oryza sativa | | Descriptor: | Alpha-amylase/subtilisin inhibitor | | Authors: | Peng, W.Y, Lin, Y.H, Huang, Y.C, Guan, H.H, Hsieh, Y.C, Chen, C.J. | | Deposit date: | 2007-07-18 | | Release date: | 2008-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Function Study of Rice Bifunctional Alpha-Amylase/Subtilisin Inhibitor from Oryza Sativa
To be Published
|
|
3LF7
 
 | |
3LIH
 
 | |
3LIG
 
 | |
3LEM
 
 | |
2QN5
 
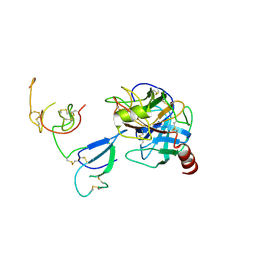 | | Crystal Structure and Functional Study of the Bowman-Birk Inhibitor from Rice Bran in Complex with Bovine Trypsin | | Descriptor: | Bowman-Birk type bran trypsin inhibitor, Cationic trypsin | | Authors: | Li, H.T, Lin, Y.H, Guan, H.H, Hsieh, Y.C, Wang, A.H.J, Chen, C.J. | | Deposit date: | 2007-07-18 | | Release date: | 2008-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure and Functional Study of the Bowman-Birk Inhibitor from Rice Bran in Complex with Bovine Trypsin
To be Published
|
|
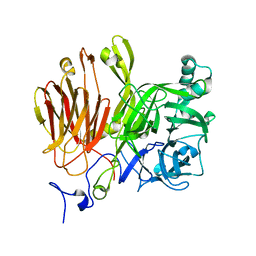
3LDR
 
 | |
4ZFH
 
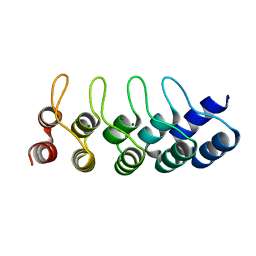 | | Crystal structure of Artificial ankyrin repeat protein_Ank(GAG)1D4 mutant -Y56A | | Descriptor: | Artificial ankyrin repeat protein_Ank(GAG)1D4 mutant -Y56A, MAGNESIUM ION | | Authors: | Chuankhayan, P, Saoin, S, Chupradit, K, Wisitponchai, T, Intachai, K, Kitidee, K, Nangola, S, Sanghiran, L.V, Hong, S.S, Boulanger, P, Tayapiwatana, C, Chen, C.J. | | Deposit date: | 2015-04-21 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of Artificial ankyrin repeat protein_Ank(GAG)1D4 mutant- Y56A
To Be Published
|
|
4QPD
 
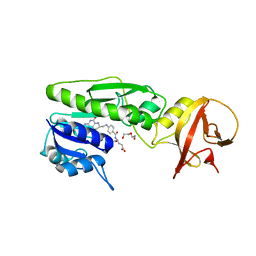 | | Crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase (wild-type) complex with tetrahydrofolate | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, 10-formyltetrahydrofolate dehydrogenase, DI(HYDROXYETHYL)ETHER | | Authors: | Lin, C.C, Chen, C.J, Fu, T.F, Chuankhayan, P, Kao, T.T, Chang, W.N. | | Deposit date: | 2014-06-23 | | Release date: | 2015-04-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the hydrolase domain of zebrafish 10-formyltetrahydrofolate dehydrogenase and its complexes reveal a complete set of key residues for hydrolysis and product inhibition.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R8V
 
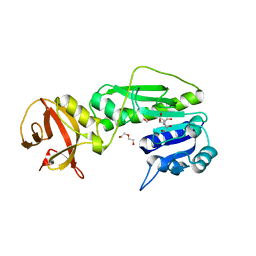 | | Crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase (wild-type) complex with formate | | Descriptor: | 10-formyltetrahydrofolate dehydrogenase, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lin, C.C, Chen, C.J, Fu, T.F, Chuankhayan, P, Kao, T.T, Chang, W.N. | | Deposit date: | 2014-09-03 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Structures of the hydrolase domain of zebrafish 10-formyltetrahydrofolate dehydrogenase and its complexes reveal a complete set of key residues for hydrolysis and product inhibition.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4TT8
 
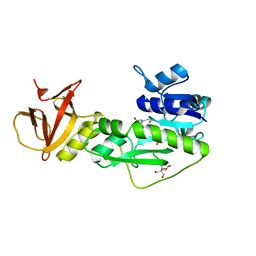 | | Crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase (wild-type) complex with 10-formyl-5,8-dideazafolate | | Descriptor: | 10-formyltetrahydrofolate dehydrogenase, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-(4-{[(2-amino-4-hydroxyquinazolin-6-yl)methyl](formyl)amino}benzoyl)-L-glutamic acid | | Authors: | Lin, C.C, Chen, C.J, Fu, T.F, Chuankhayan, P, Kao, T.T, Chang, W.N. | | Deposit date: | 2014-06-20 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structures of the hydrolase domain of zebrafish 10-formyltetrahydrofolate dehydrogenase and its complexes reveal a complete set of key residues for hydrolysis and product inhibition.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4TTS
 
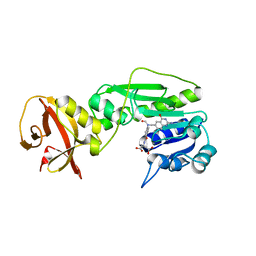 | | Crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase (Y200A) complex with 10-formyl-5,8-dideazafolate | | Descriptor: | 10-formyltetrahydrofolate dehydrogenase, N-(4-{[(2-amino-4-hydroxyquinazolin-6-yl)methyl](formyl)amino}benzoyl)-L-glutamic acid | | Authors: | Lin, C.C, Chen, C.J, Fu, T.F, Chuankhayan, P, Kao, T.T, Chang, W.N. | | Deposit date: | 2014-06-23 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the hydrolase domain of zebrafish 10-formyltetrahydrofolate dehydrogenase and its complexes reveal a complete set of key residues for hydrolysis and product inhibition.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
