5H65
 
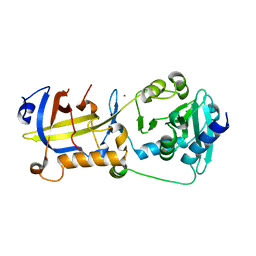 | | Crystal structure of human POT1 and TPP1 | | Descriptor: | Adrenocortical dysplasia protein homolog, Protection of telomeres protein 1, ZINC ION | | Authors: | Chen, C, Wu, J, Lei, M. | | Deposit date: | 2016-11-10 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into POT1-TPP1 interaction and POT1 C-terminal mutations in human cancer.
Nat Commun, 8, 2017
|
|
8X39
 
 | |
8X3A
 
 | |
1LYP
 
 | |
8YNJ
 
 | |
1JC6
 
 | | SOLUTION STRUCTURE OF BUNGARUS FACIATUS IX, A KUNITZ-TYPE CHYMOTRYPSIN INHIBITOR | | Descriptor: | VENOM BASIC PROTEASE INHIBITORS IX AND VIIIB | | Authors: | Chen, C, Hsu, C.H, Su, N.Y, Chiou, S.H, Wu, S.H. | | Deposit date: | 2001-06-08 | | Release date: | 2003-06-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a Kunitz-type chymotrypsin inhibitor isolated from the elapid snake Bungarus fasciatus
J.BIOL.CHEM., 276, 2001
|
|
6X5D
 
 | |
3ZZL
 
 | | Bacillus halodurans trp RNA-binding attenuation protein (TRAP): a 12- subunit assembly | | Descriptor: | TRANSCRIPTION ATTENUATION PROTEIN MTRB, TRYPTOPHAN | | Authors: | Chen, C, Smits, C, Dodson, G.G, Shevtsov, M.B, Merlino, N, Gollnick, P, Antson, A.A. | | Deposit date: | 2011-09-01 | | Release date: | 2011-10-12 | | Last modified: | 2011-10-26 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | How to Change the Oligomeric State of a Circular Protein Assembly: Switch from 11-Subunit to 12-Subunit Trap Suggests a General Mechanism
Plos One, 6, 2011
|
|
3ZZS
 
 | | Engineered 12-subunit Bacillus stearothermophilus trp RNA-binding attenuation protein (TRAP) | | Descriptor: | TRANSCRIPTION ATTENUATION PROTEIN MTRB, TRYPTOPHAN | | Authors: | Chen, C, Smits, C, Dodson, G.G, Shevtsov, M.B, Merlino, N, Gollnick, P, Antson, A.A. | | Deposit date: | 2011-09-02 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | How to Change the Oligomeric State of a Circular Protein Assembly: Switch from 11-Subunit to 12-Subunit Trap Suggests a General Mechanism
Plos One, 6, 2011
|
|
3ZZQ
 
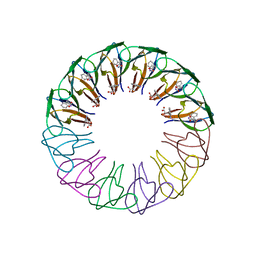 | | Engineered 12-subunit Bacillus subtilis trp RNA-binding attenuation protein (TRAP) | | Descriptor: | TRANSCRIPTION ATTENUATION PROTEIN MTRB, TRYPTOPHAN | | Authors: | Chen, C, Smits, C, Dodson, G.G, Shevtsov, M.B, Merlino, N, Gollnick, P, Antson, A.A. | | Deposit date: | 2011-09-02 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | How to Change the Oligomeric State of a Circular Protein Assembly: Switch from 11-Subunit to 12-Subunit Trap Suggests a General Mechanism
Plos One, 6, 2011
|
|
4IKA
 
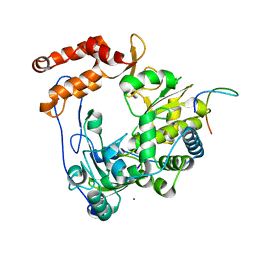 | | Crystal structure of EV71 3Dpol-VPg | | Descriptor: | 3Dpol, NICKEL (II) ION, VPg | | Authors: | Chen, C, Wang, Y.X, Lou, Z.Y. | | Deposit date: | 2012-12-25 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of enterovirus 71 RNA-dependent RNA polymerase complexed with its protein primer VPg: implication for a trans mechanism of VPg uridylylation
J.Virol., 87, 2013
|
|
2F43
 
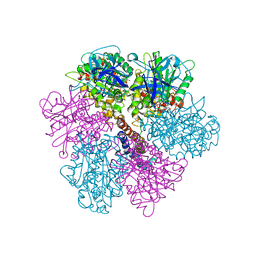 | | Rat liver F1-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase alpha chain, ... | | Authors: | Chen, C, Saxena, A.K, Simcoke, W.N, Garboczi, D.N, Pedersen, P.L, Ko, Y.H. | | Deposit date: | 2005-11-22 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mitochondrial ATP synthase: Crystal structure of the catalytic F1 unit in a vanadate-induced transition-like state and implications for mechanism.
J.Biol.Chem., 281, 2006
|
|
7RQT
 
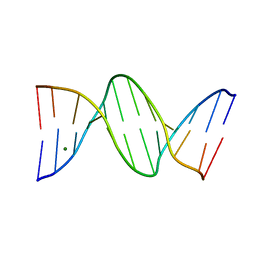 | | THE B-DNA DODECAMER With HIGH RESOLUTION | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Chen, C, Huang, Z. | | Deposit date: | 2021-08-08 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | 2'-beta-Selenium Atom on Thymidine to Control beta-Form DNA Conformation and Large Crystal Formation
Cryst.Growth Des., 2022
|
|
7V2Y
 
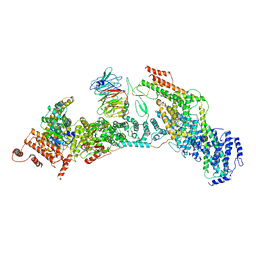 | | cryo-EM structure of yeast THO complex with Sub2 | | Descriptor: | ATP-dependent RNA helicase SUB2, Protein TEX1, THO complex subunit 2, ... | | Authors: | Chen, C, Tan, M, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2021-08-10 | | Release date: | 2022-07-27 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural and functional insights into R-loop prevention and mRNA export by budding yeast THO-Sub2 complex.
Sci Bull (Beijing), 66, 2021
|
|
7V2W
 
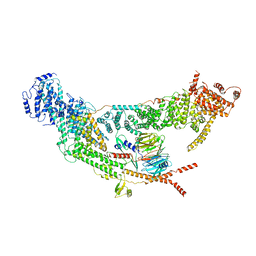 | | protomer structure from the dimer of yeast THO complex | | Descriptor: | Protein TEX1, THO complex subunit 2, THO complex subunit HPR1, ... | | Authors: | Chen, C, Tan, M, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2021-08-10 | | Release date: | 2022-07-27 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional insights into R-loop prevention and mRNA export by budding yeast THO-Sub2 complex.
Sci Bull (Beijing), 66, 2021
|
|
4N6J
 
 | | Crystal structure of human Striatin-3 coiled coil domain | | Descriptor: | Striatin-3 | | Authors: | Chen, C, Shi, Z, Zhou, Z. | | Deposit date: | 2013-10-13 | | Release date: | 2014-02-26 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Striatins contain a noncanonical coiled coil that binds protein phosphatase 2A A subunit to form a 2:2 heterotetrameric core of striatin-interacting phosphatase and kinase (STRIPAK) complex.
J.Biol.Chem., 289, 2014
|
|
3US0
 
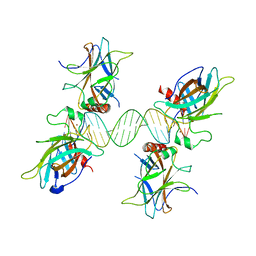 | |
3US1
 
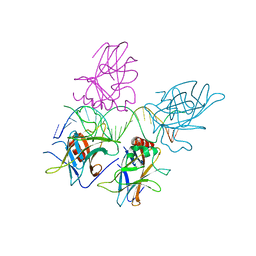 | |
3US2
 
 | |
4DML
 
 | |
4DMM
 
 | |
3HMY
 
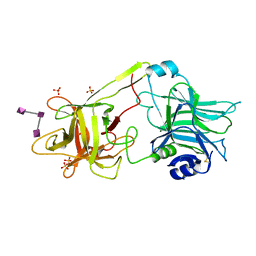 | | Crystal structure of HCR/T complexed with GT2 | | Descriptor: | GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid, SULFATE ION, ... | | Authors: | Chen, C, Fu, Z, Kim, J.-J.P, Barbieri, J.T, Baldwin, M.R. | | Deposit date: | 2009-05-29 | | Release date: | 2009-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Gangliosides as high affinity receptors for tetanus neurotoxin.
J.Biol.Chem., 284, 2009
|
|
3HN1
 
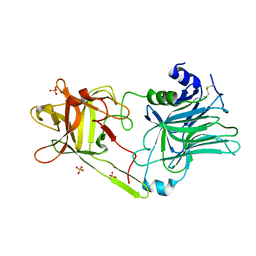 | | Crystal structure of HCR/T complexed with GT2 and lactose | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid, SULFATE ION, Tetanus toxin, ... | | Authors: | Chen, C, Fu, Z, Kim, J.-J.P, Barbieri, J.T, Baldwin, M.R. | | Deposit date: | 2009-05-29 | | Release date: | 2009-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Gangliosides as high affinity receptors for tetanus neurotoxin.
J.Biol.Chem., 284, 2009
|
|
8H61
 
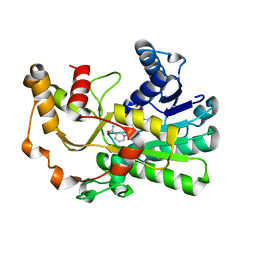 | | Ketoreductase CpKR mutant - M2 | | Descriptor: | Mutant M2 of ketoreductase CpKR, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Chen, C, Pan, J, Xu, J.H. | | Deposit date: | 2022-10-14 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Computational redesign of a robust ketoreductase for asymmetric synthesis of enantiopure diltiazem precursor.
To Be Published
|
|
3ORF
 
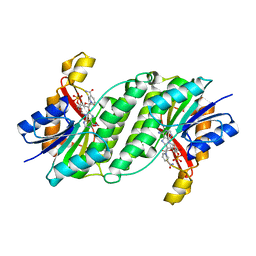 | | Crystal Structure of Dihydropteridine Reductase from Dictyostelium discoideum | | Descriptor: | Dihydropteridine reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Chen, C, Zhuang, N.N, Seo, K.H, Park, Y.S, Lee, K.H. | | Deposit date: | 2010-09-07 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural insights into the dual substrate specificities of mammalian and Dictyostelium dihydropteridine reductases toward two stereoisomers of quinonoid dihydrobiopterin
Febs Lett., 585, 2011
|
|
