3K5K
 
 | | Discovery of a 2,4-Diamino-7-aminoalkoxy-quinazoline as a Potent Inhibitor of Histone Lysine Methyltransferase, G9a | | Descriptor: | 7-[3-(dimethylamino)propoxy]-6-methoxy-2-(4-methyl-1,4-diazepan-1-yl)-N-(1-methylpiperidin-4-yl)quinazolin-4-amine, CHLORIDE ION, Histone-lysine N-methyltransferase, ... | | Authors: | Dong, A, Wasney, G.A, Liu, F, Chen, X, Allali-Hassani, A, Senisterra, G, Chau, I, Bountra, C, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Frye, S.V, Bochkarev, A, Brown, P.J, Jin, J, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-07 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of a 2,4-diamino-7-aminoalkoxyquinazoline as a potent and selective inhibitor of histone lysine methyltransferase G9a.
J.Med.Chem., 52, 2009
|
|
3N9X
 
 | | Crystal structure of Map Kinase from plasmodium berghei, PB000659.00.0 | | Descriptor: | GLYCEROL, Phosphotransferase | | Authors: | Wernimont, A.K, Hutchinson, A, Sullivan, H, MacKenzie, F, Kozieradzki, I, Chau, I, Lew, J, Senisterra, G, Artz, J, Amani, M, Cossar, D, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-31 | | Release date: | 2010-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of Map Kinase from plasmodium berghei, PB000659.00.0
To be Published
|
|
3NIE
 
 | | Crystal Structure of PF11_0147 | | Descriptor: | MAP2 kinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Wernimont, A.K, Hutchinson, A, Sullivan, H, MacKenzie, F, Kozieradzki, I, Chau, I, Lew, J, Senisterra, G, Cossar, D, Amani, M, Artz, J.D, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-06-15 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of PF11_0147
To be Published
|
|
3UR4
 
 | | Crystal structure of human WD repeat domain 5 with compound | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Dong, A, Dombrovski, L, Senisterra, G, Wernimont, A, Wasney, G.A, Allali Hassani, A, Nguyen, K.T, Smil, D, Bolshan, Y, Hajian, T, Poda, G, Chau, I, Al-Awar, R, Bountra, C, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Brown, P, Schapira, M, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-11-21 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5.
Biochem. J., 449, 2013
|
|
3RJW
 
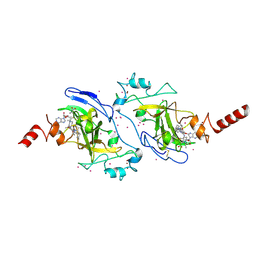 | | Crystal structure of histone lysine methyltransferase g9a with an inhibitor | | Descriptor: | 2-cyclohexyl-6-methoxy-N-[1-(1-methylethyl)piperidin-4-yl]-7-(3-pyrrolidin-1-ylpropoxy)quinazolin-4-amine, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Dong, A, Wasney, G.A, Tempel, W, Liu, F, Barsyte, D, Allali-Hassani, A, Chen, X, Chau, I, Hajian, T, Senisterra, G, Chavda, N, Arora, K, Siarheyeva, A, Kireev, D.B, Herold, J.M, Bochkarev, A, Bountra, C, Weigelt, J, Edwards, A.M, Frye, S.V, Arrowsmith, C.H, Brown, P.J, Jin, J, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-04-15 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells.
Nat.Chem.Biol., 7, 2011
|
|
5XPI
 
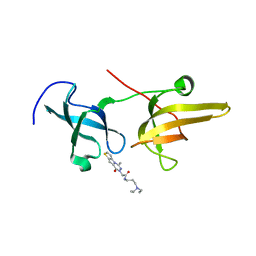 | | Structure of UHRF1 TTD in complex with NV01 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, N-[3-(diethylamino)propyl]-2-(12-methyl-9-oxidanylidene-5-thia-1,10,11-triazatricyclo[6.4.0.0^2,6]dodeca-2(6),3,7,11-tetraen-10-yl)ethanamide | | Authors: | Luo, X, Zhao, K. | | Deposit date: | 2017-06-02 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Small-Molecule Antagonists of the H3K9me3 Binding to UHRF1 Tandem Tudor Domain
SLAS Discov, 23, 2018
|
|
2WEI
 
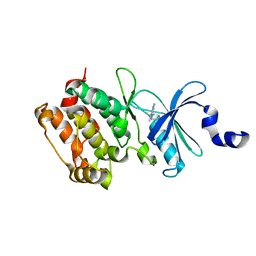 | | Crystal structure of the kinase domain of Cryptosporidium parvum calcium dependent protein kinase in complex with 3-MB-PP1 | | Descriptor: | 1-tert-butyl-3-(3-methylbenzyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, CALMODULIN-DOMAIN PROTEIN KINASE 1, PUTATIVE | | Authors: | Roos, A.K, King, O, Chaikuad, A, Zhang, C, Shokat, K.M, Wernimont, A.K, Artz, J.D, Lin, L, MacKenzie, F.I, Finerty, P.J, Vedadi, M, Schapira, M, Indarte, M, Kozieradzki, I, Pike, A.C.W, Fedorov, O, Doyle, D, Muniz, J, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, von Delft, F, Heightman, T, Hui, R. | | Deposit date: | 2009-03-31 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Cryptosporidium Parvum Kinome.
Bmc Genomics, 12, 2011
|
|
8BWU
 
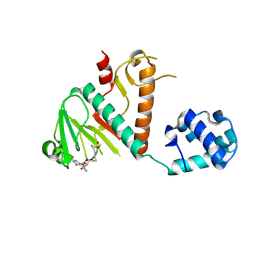 | | Crystal structure of SARS-CoV-2 nsp14 methyltransferase domain in complex with the SS148 inhibitor | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-5-cyano-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, Transcription factor ETV6,Proofreading exoribonuclease nsp14, ZINC ION | | Authors: | Konkolova, E, Klima, M, Boura, E, Jin, J, Kaniskan, H.U, Han, Y, Vedadi, M. | | Deposit date: | 2022-12-07 | | Release date: | 2023-10-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Drug Discovery in Low Data Regimes: Leveraging a Computational Pipeline for the Discovery of Novel SARS-CoV-2 Nsp14-MTase Inhibitors.
Biorxiv, 2024
|
|
6QPL
 
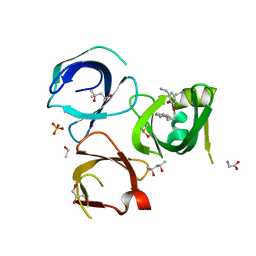 | | Crystal structure of Spindlin1 in complex with the inhibitor MS31 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Johansson, C, Krojer, T, Xiong, Y, Jin, J, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2019-02-14 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of a Potent and Selective Fragment-like Inhibitor of Methyllysine Reader Protein Spindlin 1 (SPIN1).
J.Med.Chem., 62, 2019
|
|
8GCY
 
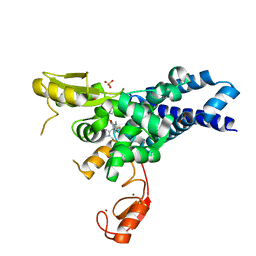 | | Co-crystal structure of CBL-B in complex with N-Aryl isoindolin-1-one inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-{3-[(1s,3R)-3-methyl-1-(4-methyl-4H-1,2,4-triazol-3-yl)cyclobutyl]phenyl}-6-{[(3S)-3-methylpiperidin-1-yl]methyl}-4-(trifluoromethyl)-2,3-dihydro-1H-isoindol-1-one, E3 ubiquitin-protein ligase CBL-B, ... | | Authors: | Kimani, S, Zeng, H, Dong, A, Li, Y, Santhakumar, V, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-03 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The co-crystal structure of Cbl-b and a small-molecule inhibitor reveals the mechanism of Cbl-b inhibition.
Commun Biol, 6, 2023
|
|
6W6D
 
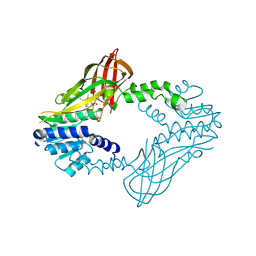 | | Crystal Structure of Human Protein arginine N-methyltransferase 6 (PRMT6) in complex with SGC6870 inhibitor | | Descriptor: | (5R)-4-(5-bromothiophene-2-carbonyl)-5-(3,5-dimethylphenyl)-7-methyl-1,3,4,5-tetrahydro-2H-1,4-benzodiazepin-2-one, Protein arginine N-methyltransferase 6, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Halabelian, L, Zeng, H, Dong, A, Jin, J, Shen, Y, Kaniskan, H.U, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A First-in-Class, Highly Selective and Cell-Active Allosteric Inhibitor of Protein Arginine Methyltransferase 6.
J.Med.Chem., 64, 2021
|
|
6XCG
 
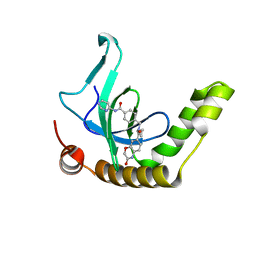 | | Histone-lysine N-methyltransferase NSD2-PWWP1 with compound UNC6934 | | Descriptor: | Histone-lysine N-methyltransferase NSD2, N-cyclopropyl-3-oxo-N-({4-[(pyrimidin-4-yl)carbamoyl]phenyl}methyl)-3,4-dihydro-2H-1,4-benzoxazine-7-carboxamide, UNKNOWN ATOM OR ION | | Authors: | Zhou, M.Q, Dong, A, Ingerman, L.A, Hanley, R.P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A chemical probe targeting the PWWP domain alters NSD2 nucleolar localization.
Nat.Chem.Biol., 18, 2022
|
|
3SMQ
 
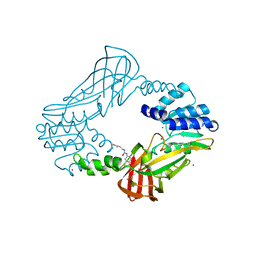 | | Crystal structure of protein arginine methyltransferase 3 | | Descriptor: | 1-(1,2,3-benzothiadiazol-6-yl)-3-[2-(cyclohex-1-en-1-yl)ethyl]urea, CHLORIDE ION, Protein arginine N-methyltransferase 3, ... | | Authors: | Dobrovetsky, E, Dong, A, Walker, J.R, Siarheyeva, A, Senisterra, G, Wasney, G.A, Smil, D, Bolshan, Y, Nguyen, K.T, Allali-Hassani, A, Hajian, T, Poda, G, Bountra, C, Weigelt, J, Edwards, A.M, Al-Awar, R, Brown, P.J, Schapira, M, Arrowsmith, C.H, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An allosteric inhibitor of protein arginine methyltransferase 3.
Structure, 20, 2012
|
|
6I8L
 
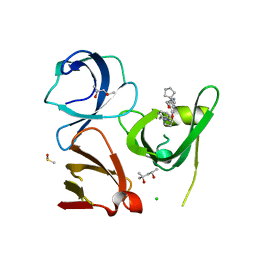 | | Crystal structure of Spindlin1 in complex with the inhibitor TD001851a | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 5'-(cyclopropylmethoxy)-6'-[3-(1,3-dihydroisoindol-2-yl)propoxy]spiro[cyclopentane-1,3'-indole]-2'-amine, ... | | Authors: | Johansson, C, Fagan, V, Brennan, P.E, Sorrell, F.J, Krojer, T, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2018-11-20 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A Chemical Probe for Tudor Domain Protein Spindlin1 to Investigate Chromatin Function.
J.Med.Chem., 62, 2019
|
|
6I8Y
 
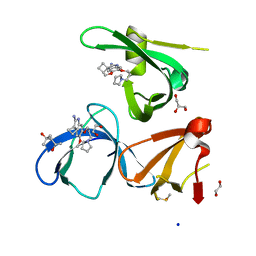 | | Crystal structure of Spindlin1 in complex with the Methyltransferase inhibitor A366 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 5'-methoxy-6'-[3-(pyrrolidin-1-yl)propoxy]spiro[cyclobutane-1,3'-indol]-2'-amine, ... | | Authors: | Srikannathasan, V, Johansson, C, Gileadi, C, Shrestha, L, Sorrell, F.J, Krojer, T, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2018-11-21 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A Chemical Probe for Tudor Domain Protein Spindlin1 to Investigate Chromatin Function.
J.Med.Chem., 62, 2019
|
|
6I8B
 
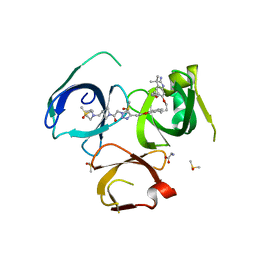 | | Crystal structure of Spindlin1 in complex with the inhibitor VinSpinIn | | Descriptor: | 2-[4-[2-[[2-[3-[2-azanyl-5-(cyclopropylmethoxy)-3,3-dimethyl-indol-6-yl]oxypropyl]-1,3-dihydroisoindol-5-yl]oxy]ethyl]-1,2,3-triazol-1-yl]-1-[4-(2-pyrrolidin-1-ylethyl)piperidin-1-yl]ethanone, DIMETHYL SULFOXIDE, GLYCINE, ... | | Authors: | Johansson, C, Fagan, V, Brennan, P.E, Sorrell, F.J, Krojer, T, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2018-11-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A Chemical Probe for Tudor Domain Protein Spindlin1 to Investigate Chromatin Function.
J.Med.Chem., 62, 2019
|
|
3SMR
 
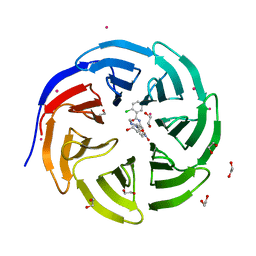 | | Crystal structure of human WD repeat domain 5 with compound | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-N-[2-(4-methylpiperazin-1-yl)-5-nitrophenyl]benzamide, UNKNOWN ATOM OR ION, ... | | Authors: | Dong, A, Dombrovski, L, Wasney, G.A, Tempel, W, Senisterra, G, Bolshan, Y, Smil, D, Nguyen, K.T, Hajian, T, Poda, G, Al-Awar, R, Bountra, C, Weigelt, J, Edwards, A.M, Brown, P.J, Schapira, M, Arrowsmith, C.H, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5.
Biochem. J., 449, 2013
|
|
7R1U
 
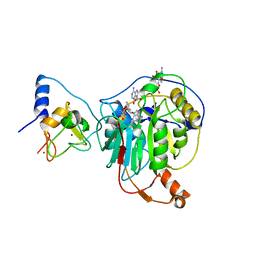 | | Crystal structure of SARS-CoV-2 nsp10/nsp16 in complex with the WZ16 inhibitor | | Descriptor: | (2S,5S)-2,6-diamino-5-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}hexanoic acid, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Klima, M, Boura, E, Li, F, Yazdi, A.K, Vedadi, M. | | Deposit date: | 2022-02-03 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10-nsp16 in complex with small molecule inhibitors, SS148 and WZ16.
Protein Sci., 31, 2022
|
|
7R1T
 
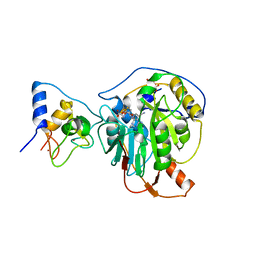 | | Crystal structure of SARS-CoV-2 nsp10/nsp16 in complex with the SS148 inhibitor | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-5-cyano-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Klima, M, Boura, E, Li, F, Yazdi, A.K, Vedadi, M. | | Deposit date: | 2022-02-03 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10-nsp16 in complex with small molecule inhibitors, SS148 and WZ16.
Protein Sci., 31, 2022
|
|
6P7I
 
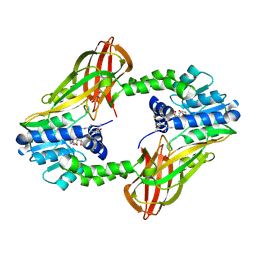 | | Crystal structure of Human PRMT6 in complex with S-Adenosyl-L-Homocysteine and YS17-117 Compound | | Descriptor: | GLYCEROL, N-[3-(4-{[(2-aminoethyl)(methyl)amino]methyl}-1H-pyrrol-3-yl)phenyl]prop-2-enamide, N-[3-(4-{[(2-aminoethyl)(methyl)amino]methyl}-1H-pyrrol-3-yl)phenyl]propanamide, ... | | Authors: | Halabelian, L, Dong, A, Zeng, H, Li, Y, Seitova, A, Hutchinson, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-05 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a First-in-Class Protein Arginine Methyltransferase 6 (PRMT6) Covalent Inhibitor
J.Med.Chem., 63, 2020
|
|
3HX4
 
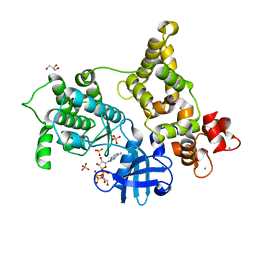 | | Crystal structure of CDPK1 of Toxoplasma gondii, TGME49_101440, in presence of calcium | | Descriptor: | CALCIUM ION, Calmodulin-domain protein kinase 1, GLYCEROL, ... | | Authors: | Wernimont, A.K, Artz, J.D, Finnerty, P, Xiao, T, He, H, MacKenzie, F, Sinestera, G, Hassani, A.A, Wasney, G, Vedadi, M, Lourido, S, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Sibley, D.L, Hui, R, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-06-19 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of apicomplexan calcium-dependent protein kinases reveal mechanism of activation by calcium.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3HZT
 
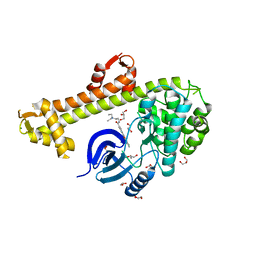 | | Crystal structure of Toxoplasma gondii CDPK3, TGME49_105860 | | Descriptor: | 5-[(E)-(5-CHLORO-2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]-N-[2-(DIETHYLAMINO)ETHYL]-2,4-DIMETHYL-1H-PYRROLE-3-CARBOXAMIDE, Calcium-dependent protein kinase 3, GLYCEROL, ... | | Authors: | Wernimont, A.K, Artz, J.D, Finnerty, P, Wasney, G, Allali-Hassani, A, Vedadi, M, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-06-24 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of apicomplexan calcium-dependent protein kinases reveal mechanism of activation by calcium.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3IGO
 
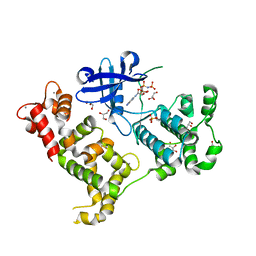 | | Crystal structure of Cryptosporidium parvum CDPK1, cgd3_920 | | Descriptor: | CALCIUM ION, Calmodulin-domain protein kinase 1, GLYCEROL, ... | | Authors: | Wernimont, A.K, Artz, J.D, Finnerty, P, Amani, M, Allali-Hassanali, A, Vedadi, M, Tempel, W, MacKenzie, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Bochkarev, A, Hui, R, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-28 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of apicomplexan calcium-dependent protein kinases reveal mechanism of activation by calcium.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KU2
 
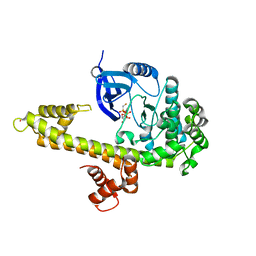 | | Crystal Structure of inactivated form of CDPK1 from toxoplasma gondii, TGME49.101440 | | Descriptor: | Calmodulin-domain protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, UNKNOWN ATOM OR ION | | Authors: | Wernimont, A.K, Artz, J.D, Finnerty, P, Xiao, T, He, H, Mackenzie, F, Sinestera, G, Hassani, A.A, Wasney, G, Vedadi, M, Lourido, S, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Sibley, D.L, Hui, R, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-26 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of apicomplexan calcium-dependent protein kinases reveal mechanism of activation by calcium.
Nat.Struct.Mol.Biol., 17, 2010
|
|
7MDN
 
 | | Histone-lysine N-methyltransferase NSD2-PWWP1 with compound MRT10241866a | | Descriptor: | Histone-lysine N-methyltransferase NSD2, UNKNOWN ATOM OR ION, ~{N}-cyclopropyl-3-oxidanylidene-~{N}-(thiophen-2-ylmethyl)-4~{H}-1,4-benzoxazine-7-carboxamide | | Authors: | Lei, M, Freitas, R.F, Dong, A, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-05 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | A chemical probe targeting the PWWP domain alters NSD2 nucleolar localization.
Nat.Chem.Biol., 18, 2022
|
|
