1QGQ
 
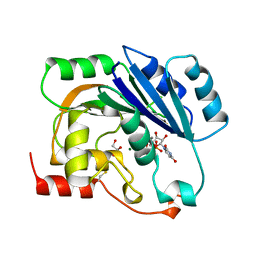 | | UDP-MANGANESE COMPLEX OF SPSA FROM BACILLUS SUBTILIS | | Descriptor: | GLYCEROL, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Charnock, S.J. | | Deposit date: | 1999-05-04 | | Release date: | 2000-05-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the nucleotide-diphospho-sugar transferase, SpsA from Bacillus subtilis, in native and nucleotide-complexed forms.
Biochemistry, 38, 1999
|
|
1QG8
 
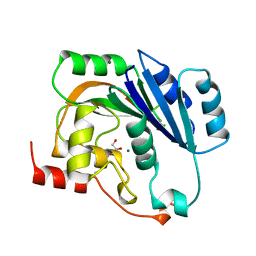 | | NATIVE (MAGNESIUM-CONTAINING) SPSA FROM BACILLUS SUBTILIS | | Descriptor: | GLYCEROL, MAGNESIUM ION, PROTEIN (SPORE COAT POLYSACCHARIDE BIOSYNTHESIS PROTEIN SPSA) | | Authors: | Charnock, S.J. | | Deposit date: | 1999-04-21 | | Release date: | 2000-04-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the nucleotide-diphospho-sugar transferase, SpsA from Bacillus subtilis, in native and nucleotide-complexed forms.
Biochemistry, 38, 1999
|
|
1QGS
 
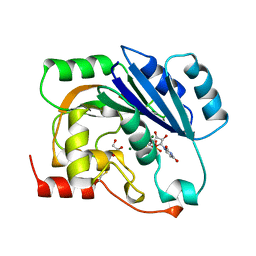 | | UDP-MAGNESIUM COMPLEX OF SPSA FROM BACILLUS SUBTILIS | | Descriptor: | GLYCEROL, MAGNESIUM ION, PROTEIN (SPORE COAT POLYSACCHARIDE BIOSYNTHESIS PROTEIN SPSA), ... | | Authors: | Charnock, S.J. | | Deposit date: | 1999-05-04 | | Release date: | 2000-05-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the nucleotide-diphospho-sugar transferase, SpsA from Bacillus subtilis, in native and nucleotide-complexed forms.
Biochemistry, 38, 1999
|
|
1GXN
 
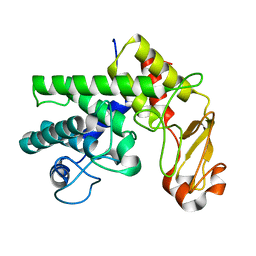 | | Family 10 polysaccharide lyase from Cellvibrio cellulosa | | Descriptor: | PECTATE LYASE | | Authors: | Charnock, S.J, Brown, I.E, Turkenburg, J.P, Black, G.W, Davies, G.J. | | Deposit date: | 2002-04-08 | | Release date: | 2002-10-04 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Convergent Evolution Sheds Light on the Anti-Beta-Elimination Mechanism Common to Family 1 and 10 Polysaccharide Lyases
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1GXM
 
 | | Family 10 polysaccharide lyase from Cellvibrio cellulosa | | Descriptor: | GLYCEROL, PECTATE LYASE | | Authors: | Charnock, S.J, Brown, I.E, Turkenburg, J.P, Black, G.W, Davies, G.J. | | Deposit date: | 2002-04-08 | | Release date: | 2002-10-04 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Convergent Evolution Sheds Light on the Anti-Beta-Elimination Mechanism Common to Family 1 and 10 Polysaccharide Lyases
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1GXO
 
 | | Mutant D189A of Family 10 polysaccharide lyase from Cellvibrio cellulosa in complex with trigalaturonic acid | | Descriptor: | CALCIUM ION, PECTATE LYASE, alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid | | Authors: | Charnock, S.J, Brown, I.E, Turkenburg, J.P, Black, G.W, Davies, G.J. | | Deposit date: | 2002-04-08 | | Release date: | 2002-10-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Convergent Evolution Sheds Light on the Anti-Beta-Elimination Mechanism Common to Family 1 and 10 Polysaccharide Lyases
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1GWK
 
 | | Carbohydrate binding module family29 | | Descriptor: | NON-CATALYTIC PROTEIN 1 | | Authors: | Charnock, S.J, Nurizzo, D, Davies, G.J. | | Deposit date: | 2002-03-19 | | Release date: | 2003-03-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Promiscuity in Ligand-Binding: The Three-Dimensional Structure of a Piromyces Carbohydrate-Binding Module,Cbm29-2,in Complex with Cello- and Mannohexaose
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1GWL
 
 | | Carbohydrate binding module family29 complexed with mannohexaose | | Descriptor: | NON-CATALYTIC PROTEIN 1, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Charnock, S.J, Nurizzo, D, Davies, G.J. | | Deposit date: | 2002-03-19 | | Release date: | 2003-03-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Promiscuity in Ligand-Binding: The Three-Dimensional Structure of a Piromyces Carbohydrate-Binding Module,Cbm29-2,in Complex with Cello- and Mannohexaose
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1GWM
 
 | | Carbohydrate binding module family29 complexed with glucohexaose | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, NON-CATALYTIC PROTEIN 1, ... | | Authors: | Charnock, S.J, Nurizzo, D, Davies, G.J. | | Deposit date: | 2002-03-19 | | Release date: | 2003-03-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Promiscuity in Ligand-Binding: The Three-Dimensional Structure of a Piromyces Carbohydrate-Binding Module,Cbm29-2,in Complex with Cello- and Mannohexaose
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1DYO
 
 | | Xylan-Binding Domain from CBM 22, formally x6b domain | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE Y | | Authors: | Davies, G.J, Charnock, S.J, Gilbert, H.J, Fontes, C.M.G.A. | | Deposit date: | 2000-02-03 | | Release date: | 2000-07-04 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The X6 Thermostabilising Domains of Xylanases are Carbohydrate Binding Modules: Structure and Biochemistry of the Clostridium Thermocellum X6B Domain
Biochemistry, 39, 2000
|
|
2C3F
 
 | | The structure of a group A streptococcal phage-encoded tail-fibre showing hyaluronan lyase activity. | | Descriptor: | HYALURONIDASE, PHAGE ASSOCIATED, SODIUM ION | | Authors: | Taylor, E.J, Smith, N.L, Linsay, A.-M, Charnock, S.J, Turkenburg, J.P, Dodson, E.J, Davies, G.J, Black, G.W. | | Deposit date: | 2005-10-06 | | Release date: | 2005-11-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure of a Group a Streptococcal Phage-Encoded Virulence Factor Reveals Catalytically Active Triple-Stranded Beta-Helix
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2WCO
 
 | | Structures of the Streptomyces coelicolor A3(2) Hyaluronan Lyase in Complex with Oligosaccharide Substrates and an Inhibitor | | Descriptor: | 4-deoxy-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, FORMIC ACID, GLYCEROL, ... | | Authors: | Elmabrouk, Z.H, Taylor, E.J, Vincent, F, Smith, N.L, Zhang, M, Charnock, S.J, Turkenburg, J.P, Davies, G.J, Black, G.W. | | Deposit date: | 2009-03-12 | | Release date: | 2010-08-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structures of a Family 8 Polysaccharide Lyase Reveal Open and Highly Occluded Substrate-Binding Cleft Conformations.
Proteins, 79, 2011
|
|
1H6Y
 
 | | The role of conserved amino acids in the cleft of the C-terminal family 22 carbohydrate binding module of Clostridium thermocellum Xyn10B in ligand binding | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE Y | | Authors: | Xie, H, Bolam, D.N, Charnock, S.J, Davies, G.J, Williamson, M.P, Simpson, P.J, Fontes, C.M.G.A, Ferreira, L.M.A, Gilbert, H.J. | | Deposit date: | 2001-06-29 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Clostridium Thermocellum Xyn10B Carbohydrate-Binding Module 22-2: The Role of Conserved Amino Acids in Ligand Binding
Biochemistry, 40, 2001
|
|
1US2
 
 | | Xylanase10C (mutant E385A) from Cellvibrio japonicus in complex with xylopentaose | | Descriptor: | ENDO-BETA-1,4-XYLANASE, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Pell, G, Szabo, L, Charnock, S.J, Xie, H, Gloster, T.M, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2003-11-17 | | Release date: | 2003-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biochemical Analysis of Cellvibrio Japonicus Xylanase 10C: How Variation in Substrate-Binding Cleft Influences the Catalytic Profile of Family Gh-10 Xylanases
J.Biol.Chem., 279, 2004
|
|
1US3
 
 | | Native xylanase10C from Cellvibrio japonicus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ENDO-BETA-1,4-XYLANASE PRECURSOR, GLYCEROL, ... | | Authors: | Pell, G, Szabo, L, Charnock, S.J, Xie, H, Gloster, T.M, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2003-11-17 | | Release date: | 2003-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biochemical Analysis of Cellvibrio Japonicus Xylanase 10C: How Variation in Substrate-Binding Cleft Influences the Catalytic Profile of Family Gh-10 Xylanases
J.Biol.Chem., 279, 2004
|
|
1H6X
 
 | | The role of conserved amino acids in the cleft of the C-terminal family 22 carbohydrate binding module of Clostridium thermocellum Xyn10B in ligand binding | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE Y | | Authors: | Xie, H, Bolam, D.N, Charnock, S.J, Davies, G.J, Williamson, M.P, Simpson, P.J, Fontes, C.M.G.A, Ferreira, L.M.A, Gilbert, H.J. | | Deposit date: | 2001-06-29 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Clostridium Thermocellum Xyn10B Carbohydrate-Binding Module 22-2: The Role of Conserved Amino Acids in Ligand Binding
Biochemistry, 40, 2001
|
|
1ODS
 
 | | Cephalosporin C deacetylase from Bacillus subtilis | | Descriptor: | CEPHALOSPORIN C DEACETYLASE, CHLORIDE ION, MAGNESIUM ION | | Authors: | Vincent, F, Charnock, S.J, Verschueren, K.H.G, Turkenburg, J.P, Scott, D.J, Offen, W.A, Roberts, S, Pell, G, Gilbert, H.J, Brannigan, J.A, Davies, G.J. | | Deposit date: | 2003-02-20 | | Release date: | 2003-07-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multifunctional Xylooligosaccharide/Cephalosporin C Deacetylase Revealed by the Hexameric Structure of the Bacillus Subtilis Enzyme at 1.9A Resolution
J.Mol.Biol., 330, 2003
|
|
1ODT
 
 | | cephalosporin C deacetylase mutated, in complex with acetate | | Descriptor: | ACETATE ION, CEPHALOSPORIN C DEACETYLASE | | Authors: | Vincent, F, Charnock, S.J, Verschueren, K.H.G, Turkenburg, J.P, Scott, D.J, Offen, W.A, Roberts, S, Pell, G, Gilbert, H.J, Brannigan, J.A, Davies, G.J. | | Deposit date: | 2003-02-20 | | Release date: | 2003-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Multifunctional Xylooligosaccharide/Cephalosporin C Deacetylase Revealed by the Hexameric Structure of the Bacillus Subtilis Enzyme at 1.9A Resolution
J.Mol.Biol., 330, 2003
|
|
1E0W
 
 | | Xylanase 10A from Sreptomyces lividans. native structure at 1.2 angstrom resolution | | Descriptor: | ENDO-1,4-BETA-XYLANASE A | | Authors: | Ducros, V, Charnock, S.J, Derewenda, U, Derewenda, Z.S, Dauter, Z, Dupont, C, Shareck, F, Morosoli, R, Kluepfel, D, Davies, G.J. | | Deposit date: | 2000-04-10 | | Release date: | 2001-04-05 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Substrate Specificity in Glycoside Hydrolase Family 10. Structural and Kinetic Analysis of the Streptomyces Lividans Xylanase 10A
J.Biol.Chem., 275, 2000
|
|
1E0V
 
 | | Xylanase 10A from Sreptomyces lividans. cellobiosyl-enzyme intermediate at 1.7 A | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, beta-D-glucopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-glucopyranose | | Authors: | Ducros, V, Charnock, S.J, Derewenda, U, Derewenda, Z.S, Dauter, Z, Dupont, C, Shareck, F, Morosoli, R, Kluepfel, D, Davies, G.J. | | Deposit date: | 2000-04-10 | | Release date: | 2001-04-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate Specificity in Glycoside Hydrolase Family 10. Structural and Kinetic Analysis of the Streptomyces Lividans Xylanase 10A
J.Biol.Chem., 275, 2000
|
|
1H7Q
 
 | | dTDP-MANGANESE COMPLEX OF SPSA FROM BACILLUS SUBTILIS | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, SPORE COAT POLYSACCHARIDE BIOSYNTHESIS PROTEIN SPSA, ... | | Authors: | Tarbouriech, N, Charnock, S.J, Davies, G.J. | | Deposit date: | 2001-07-09 | | Release date: | 2001-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-Dimensional Structures of the Mn and Mg Dtdp Complexes of the Family Gt-2 Glycosyltransferase Spsa: A Comparison with Related Ndp-Sugar Glycosyltransferases.
J.Mol.Biol., 314, 2001
|
|
1H7L
 
 | | dTDP-MAGNESIUM COMPLEX OF SPSA FROM BACILLUS SUBTILIS | | Descriptor: | MAGNESIUM ION, SPORE COAT POLYSACCHARIDE BIOSYNTHESIS PROTEIN SPSA, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Tarbouriech, N, Charnock, S.J, Davies, G.J. | | Deposit date: | 2001-07-09 | | Release date: | 2001-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Three-Dimensional Structures of the Mn and Mg Dtdp Complexes of the Family Gt-2 Glycosyltransferase Spsa: A Comparison with Related Ndp-Sugar Glycosyltransferases.
J.Mol.Biol., 314, 2001
|
|
1E0X
 
 | | XYLANASE 10A FROM SREPTOMYCES LIVIDANS. XYLOBIOSYL-ENZYME INTERMEDIATE AT 1.65 A | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, GLYCEROL, beta-D-xylopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-xylopyranose | | Authors: | Ducros, V, Charnock, S.J, Derewenda, U, Derewenda, Z.S, Dauter, Z, Dupont, C, Shareck, F, Morosoli, R, Kluepfel, D, Davies, G.J. | | Deposit date: | 2000-04-10 | | Release date: | 2001-04-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Substrate Specificity in Glycoside Hydrolase Family 10. Structural and Kinetic Analysis of the Streptomyces Lividans Xylanase 10A
J.Biol.Chem., 275, 2000
|
|
1GKL
 
 | | S954A mutant of the feruloyl esterase module from clostridium thermocellum complexed with ferulic acid | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, ACETATE ION, CADMIUM ION, ... | | Authors: | Prates, J.A.M, Tarbouriech, N, Charnock, S.J, Fontes, C.M.G.A, Ferreira, L.M.A, Davies, G.J. | | Deposit date: | 2001-08-15 | | Release date: | 2001-12-13 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The structure of the feruloyl esterase module of xylanase 10B from Clostridium thermocellum provides insights into substrate recognition.
Structure, 9, 2001
|
|
1GKK
 
 | | Feruloyl esterase domain of XynY from clostridium thermocellum | | Descriptor: | CADMIUM ION, ENDO-1,4-BETA-XYLANASE Y, GLYCEROL | | Authors: | Prates, J.A.M, Tarbouriech, N, Charnock, S.J, Fontes, C.M.G.A, Ferreira, L.M.A, Davies, G.J. | | Deposit date: | 2001-08-15 | | Release date: | 2001-12-13 | | Last modified: | 2011-09-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure of the Feruloyl Esterase Module of Xylanase 10B from Clostridium Thermocellum Provides Insights Into Substrate Recognition
Structure, 9, 2001
|
|
