1Z6I
 
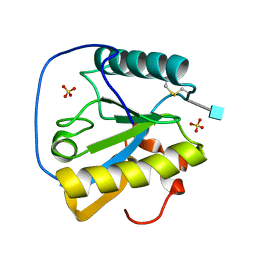 | | Crystal structure of the ectodomain of Drosophila transmembrane receptor PGRP-LCa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Peptidoglycan-recognition protein-LC, SULFATE ION | | Authors: | Chang, C.-I, Ihara, K, Chelliah, Y, Mengin-Lecreulx, D, Wakatsuki, S, Deisenhofer, J. | | Deposit date: | 2005-03-22 | | Release date: | 2005-07-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the ectodomain of Drosophila peptidoglycan-recognition protein LCa suggests a molecular mechanism for pattern recognition
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1S2J
 
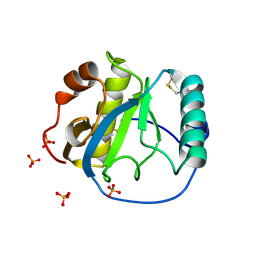 | | Crystal structure of the Drosophila pattern-recognition receptor PGRP-SA | | Descriptor: | PHOSPHATE ION, Peptidoglycan recognition protein SA CG11709-PA | | Authors: | Chang, C.-I, Pili-Floury, S, Chelliah, Y, Lemaitre, B, Mengin-Lecreulx, D, Deisenhofer, J. | | Deposit date: | 2004-01-08 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Drosophila pattern recognition receptor contains a peptidoglycan docking groove and unusual l,d-carboxypeptidase activity.
PLOS BIOL., 2, 2004
|
|
4YPN
 
 | |
5E7S
 
 | |
4YPM
 
 | |
4XH6
 
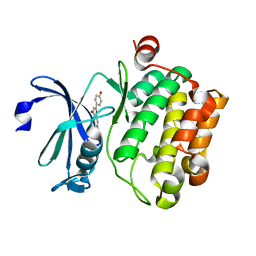 | | Crystal structure of proto-oncogene kinase Pim1 bound to hispidulin | | Descriptor: | 5,7-dihydroxy-2-(4-hydroxyphenyl)-6-methoxy-4H-chromen-4-one, Serine/threonine-protein kinase pim-1 | | Authors: | Su, M.-Y, Chang, C.-I. | | Deposit date: | 2015-01-05 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Total Synthesis of Hispidulin and the Structural Basis for Its Inhibition of Proto-oncogene Kinase Pim-1
J.Nat.Prod., 78, 2015
|
|
4YPL
 
 | | Crystal structure of a hexameric LonA protease bound to three ADPs | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease, N-[(1R)-1-(dihydroxyboranyl)-2-phenylethyl]-Nalpha-(pyrazin-2-ylcarbonyl)-L-phenylalaninamide | | Authors: | Lin, C.-C, Chang, C.-I. | | Deposit date: | 2015-03-13 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structural Insights into the Allosteric Operation of the Lon AAA+ Protease
Structure, 24, 2016
|
|
1LEW
 
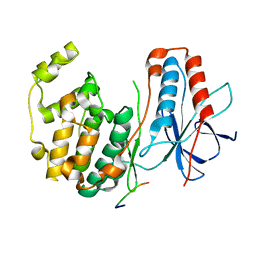 | | CRYSTAL STRUCTURE OF MAP KINASE P38 COMPLEXED TO THE DOCKING SITE ON ITS NUCLEAR SUBSTRATE MEF2A | | Descriptor: | Mitogen-activated protein kinase 14, Myocyte-specific enhancer factor 2A | | Authors: | Chang, C.-I, Xu, B.-E, Akella, R, Cobb, M.H, Goldsmith, E.J. | | Deposit date: | 2002-04-10 | | Release date: | 2002-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of MAP kinase p38 complexed to the docking sites on its nuclear substrate MEF2A and activator MKK3b.
Mol.Cell, 9, 2002
|
|
1LEZ
 
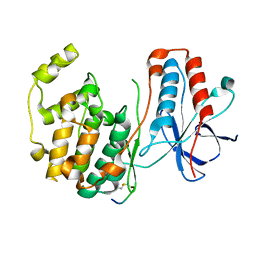 | | CRYSTAL STRUCTURE OF MAP KINASE P38 COMPLEXED TO THE DOCKING SITE ON ITS ACTIVATOR MKK3B | | Descriptor: | MAP kinase kinase 3b, MITOGEN-ACTIVATED PROTEIN KINASE 14 | | Authors: | Chang, C.-I, Xu, B.-E, Akella, R, Cobb, M.H, Goldsmith, E.J. | | Deposit date: | 2002-04-10 | | Release date: | 2002-07-10 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of MAP kinase p38 complexed to the docking sites on its nuclear substrate MEF2A and activator MKK3b.
Mol.Cell, 9, 2002
|
|
8IKR
 
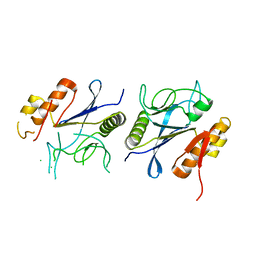 | | Crystal structure of DpaA | | Descriptor: | YkuD domain-containing protein | | Authors: | Wang, H.-J, Hsieh, K.-Y, Lee, S.-H, Chang, C.-I. | | Deposit date: | 2023-03-01 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the hydrolytic activity of the transpeptidase-like protein DpaA to detach Braun's lipoprotein from peptidoglycan.
Mbio, 14, 2023
|
|
4R8F
 
 | |
7CRA
 
 | |
7CR9
 
 | |
