4JDR
 
 | | Dihydrolipoamide dehydrogenase of pyruvate dehydrogenase from escherichia coli | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Chandrasekhar, K, Arjunan, P, Furey, W. | | Deposit date: | 2013-02-25 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight to the Interaction of the Dihydrolipoamide Acetyltransferase (E2) Core with the Peripheral Components in the Escherichia coli Pyruvate Dehydrogenase Complex via Multifaceted Structural Approaches.
J.Biol.Chem., 288, 2013
|
|
1QC9
 
 | |
4N72
 
 | |
2G67
 
 | |
1RP7
 
 | | E. COLI PYRUVATE DEHYDROGENASE INHIBITOR COMPLEX | | Descriptor: | 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, MAGNESIUM ION, Pyruvate dehydrogenase E1 component | | Authors: | Arjunan, P, Chandrasekhar, K, Furey, W. | | Deposit date: | 2003-12-03 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural Determinants of Enzyme Binding Affinity: The E1 Component of Pyruvate Dehydrogenase from Escherichia coli in Complex with the Inhibitor Thiamin Thiazolone Diphosphate.
Biochemistry, 43, 2004
|
|
2G25
 
 | | E. Coli Pyruvate Dehydrogenase Phosphonolactylthiamin Diphosphate Complex | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-{(1S)-1-HYDROXY-1-[(R)-HYDROXY(METHOXY)PHOSPHORYL]ETHYL}-5-(2-{[(S)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Furey, W, Arjunan, P, Chandrasekhar, K. | | Deposit date: | 2006-02-15 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Thiamin-bound, Pre-decarboxylation Reaction Intermediate Analogue in the Pyruvate Dehydrogenase E1 Subunit Induces Large Scale Disorder-to-Order Transformations in the Enzyme and Reveals Novel Structural Features in the Covalently Bound Adduct.
J.Biol.Chem., 281, 2006
|
|
2G28
 
 | | E. Coli Pyruvate Dehydrogenase H407A variant Phosphonolactylthiamin Diphosphate Complex | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-{(1S)-1-HYDROXY-1-[(R)-HYDROXY(METHOXY)PHOSPHORYL]ETHYL}-5-(2-{[(S)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, MAGNESIUM ION, Pyruvate dehydrogenase E1 component | | Authors: | Furey, W, Arjunan, P, Chandrasekhar, K. | | Deposit date: | 2006-02-15 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Thiamin-bound, Pre-decarboxylation Reaction Intermediate Analogue in the Pyruvate Dehydrogenase E1 Subunit Induces Large Scale Disorder-to-Order Transformations in the Enzyme and Reveals Novel Structural Features in the Covalently Bound Adduct.
J.Biol.Chem., 281, 2006
|
|
3LPL
 
 | | E. coli pyruvate dehydrogenase complex E1 component E571A mutant | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Furey, W. | | Deposit date: | 2010-02-05 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Communication between thiamin cofactors in the Escherichia coli pyruvate dehydrogenase complex E1 component active centers: evidence for a "direct pathway" between the 4'-aminopyrimidine N1' atoms.
J.Biol.Chem., 285, 2010
|
|
4QOY
 
 | |
8LDH
 
 | |
3LQ2
 
 | | E. coli pyruvate dehydrogenase complex E1 E235A mutant with low TDP concentration | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Furey, W. | | Deposit date: | 2010-02-08 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Communication between thiamin cofactors in the Escherichia coli pyruvate dehydrogenase complex E1 component active centers: evidence for a "direct pathway" between the 4'-aminopyrimidine N1' atoms.
J.Biol.Chem., 285, 2010
|
|
3LQ4
 
 | | E. coli pyruvate dehydrogenase complex E1 E235A mutant with high TDP concentration | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Furey, W. | | Deposit date: | 2010-02-08 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Communication between thiamin cofactors in the Escherichia coli pyruvate dehydrogenase complex E1 component active centers: evidence for a "direct pathway" between the 4'-aminopyrimidine N1' atoms.
J.Biol.Chem., 285, 2010
|
|
6LDH
 
 | |
1LDM
 
 | |
1L8A
 
 | | E. COLI PYRUVATE DEHYDROGENASE | | Descriptor: | MAGNESIUM ION, Pyruvate dehydrogenase E1 component, THIAMINE DIPHOSPHATE | | Authors: | Furey, W, Arjunan, P. | | Deposit date: | 2002-03-19 | | Release date: | 2002-07-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the pyruvate dehydrogenase multienzyme complex E1 component from Escherichia coli at 1.85 A resolution.
Biochemistry, 41, 2002
|
|
3LDH
 
 | | A comparison of the structures of apo dogfish m4 lactate dehydrogenase and its ternary complexes | | Descriptor: | LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PYRUVIC ACID | | Authors: | White, J.L, Hackert, M.L, Buehner, M, Adams, M.J, Ford, G.C, Lentzjunior, P.J, Smiley, I.E, Steindel, S.J, Rossmann, M.G. | | Deposit date: | 1974-06-06 | | Release date: | 1977-04-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A comparison of the structures of apo dogfish M4 lactate dehydrogenase and its ternary complexes.
J.Mol.Biol., 102, 1976
|
|
1CKQ
 
 | |
1XOB
 
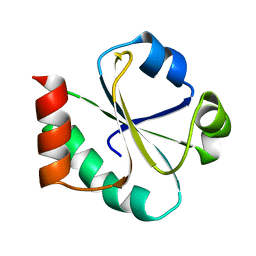 | | THIOREDOXIN (REDUCED DITHIO FORM), NMR, 20 STRUCTURES | | Descriptor: | THIOREDOXIN | | Authors: | Jeng, M.-F, Campbell, A.P, Begley, T, Holmgren, A, Case, D.A, Wright, P.E, Dyson, H.J. | | Deposit date: | 1995-11-28 | | Release date: | 1996-06-10 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structures of oxidized and reduced Escherichia coli thioredoxin.
Structure, 2, 1994
|
|
1XOA
 
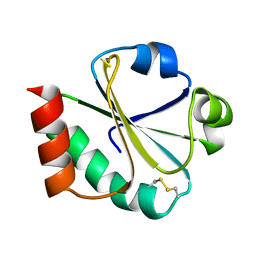 | | THIOREDOXIN (OXIDIZED DISULFIDE FORM), NMR, 20 STRUCTURES | | Descriptor: | THIOREDOXIN | | Authors: | Jeng, M.-F, Campbell, A.P, Begley, T, Holmgren, A, Case, D.A, Wright, P.E, Dyson, H.J. | | Deposit date: | 1995-11-28 | | Release date: | 1996-06-10 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structures of oxidized and reduced Escherichia coli thioredoxin.
Structure, 2, 1994
|
|
2IEA
 
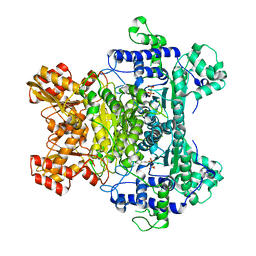 | | E. coli pyruvate dehydrogenase | | Descriptor: | MAGNESIUM ION, Pyruvate dehydrogenase E1 component, THIAMINE DIPHOSPHATE | | Authors: | Furey, W, Arjunan, P. | | Deposit date: | 2006-09-18 | | Release date: | 2006-10-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the pyruvate dehydrogenase multienzyme complex E1 component from Escherichia coli at 1.85 A resolution.
Biochemistry, 41, 2002
|
|
2QTC
 
 | | E. coli Pyruvate dehydrogenase E1 component E401K mutant with phosphonolactylthiamin diphosphate | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-{(1S)-1-HYDROXY-1-[(R)-HYDROXY(METHOXY)PHOSPHORYL]ETHYL}-5-(2-{[(S)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, MAGNESIUM ION, Pyruvate dehydrogenase E1 component | | Authors: | Furey, W, Arjunan, P, Chandrasekhar, K. | | Deposit date: | 2007-08-01 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A Dynamic Loop at the Active Center of the Escherichia coli Pyruvate Dehydrogenase Complex E1 Component Modulates Substrate Utilization and Chemical Communication with the E2 Component
J.Biol.Chem., 282, 2007
|
|
2QTA
 
 | |
1ERI
 
 | | X-RAY STRUCTURE OF THE DNA-ECO RI ENDONUCLEASE-DNA RECOGNITION COMPLEX: THE RECOGNITION NETWORK AND THE INTEGRATION OF RECOGNITION AND CLEAVAGE | | Descriptor: | DNA (5'-D(*TP*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), PROTEIN (ECO RI ENDONUCLEASE (E.C.3.1.21.4)) | | Authors: | Kim, Y, Grable, J.C, Love, R, Greene, P.J, Rosenberg, J.M. | | Deposit date: | 1994-05-18 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Refinement of Eco RI endonuclease crystal structure: a revised protein chain tracing.
Science, 249, 1990
|
|
