5EPD
 
 | | Crystal structure of Glycerol Trinitrate Reductase XdpB from Agrobacterium sp. R89-1 (Apo form) | | Descriptor: | Glycerol trinitrate reductase | | Authors: | Kolenko, P, Zahradnik, J, Zuskova, I, Cerny, J, Palyzova, A, Kyslikova, E, Schneider, B. | | Deposit date: | 2015-11-11 | | Release date: | 2016-11-23 | | Last modified: | 2019-02-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of XdpB, the bacterial old yellow enzyme, in an FMN-free form.
PLoS ONE, 13, 2018
|
|
5EH1
 
 | | Crystal structure of the extracellular part of receptor 2 of human interferon gamma | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYSTEINE, GLYCEROL, ... | | Authors: | Kolenko, P, Mikulecky, P, Zahradnik, J, Dohnalek, J, Koval, T, Cerny, J, Necasova, I, Schneider, B. | | Deposit date: | 2015-10-27 | | Release date: | 2016-08-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human interferon-gamma receptor 2 reveals the structural basis for receptor specificity.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4OC5
 
 | | X-ray structure of of human glutamate carboxypeptidase II (GCPII) in a complex with CHIBzL, a urea-based inhibitor N~2~-{[(S)-carboxy(4-hydroxyphenyl)methyl]carbamoyl}-N~6~-(4-iodobenzoyl)-L-lysine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Pavlicek, J, Ptacek, J, Cerny, J, Byun, Y, Skultetyova, L, Pomper, M, Lubkowski, J, Barinka, C. | | Deposit date: | 2014-01-08 | | Release date: | 2014-05-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural characterization of P1'-diversified urea-based inhibitors of glutamate carboxypeptidase II.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4OC1
 
 | | X-ray structure of of human glutamate carboxypeptidase II (GCPII) in a complex with COIBzL, a urea-based inhibitor (2S)-2-[({(1S)-1-carboxy-2-[(2S)-oxiran-2-yl]ethyl}carbamoyl)amino]-6-[(4-iodobenzoyl)amino]hexanoic acid | | Descriptor: | (2S)-2-[({(1S)-1-carboxy-2-[(2S)-oxiran-2-yl]ethyl}carbamoyl)amino]-6-[(4-iodobenzoyl)amino]hexanoic acid (non-preferred name), 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pavlicek, J, Ptacek, J, Cerny, J, Byun, Y, Skultetyova, L, Pomper, M, Lubkowski, J, Barinka, C. | | Deposit date: | 2014-01-08 | | Release date: | 2014-05-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural characterization of P1'-diversified urea-based inhibitors of glutamate carboxypeptidase II.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4OC2
 
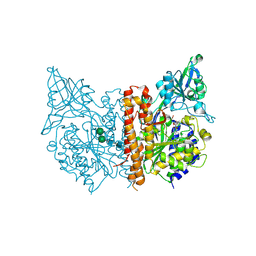 | | X-ray structure of of human glutamate carboxypeptidase II (GCPII) in a complex with CEIBzL, a urea-based inhibitor N~2~-{[(1S)-1-carboxybut-3-yn-1-yl]carbamoyl}-N~6~-(4-iodobenzoyl)-L-lysine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pavlicek, J, Ptacek, J, Cerny, J, Byun, Y, Skultetyova, L, Pomper, M, Lubkowski, J, Barinka, C. | | Deposit date: | 2014-01-08 | | Release date: | 2014-05-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural characterization of P1'-diversified urea-based inhibitors of glutamate carboxypeptidase II.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4OC0
 
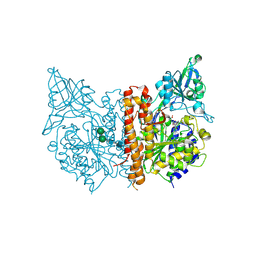 | | X-ray structure of of human glutamate carboxypeptidase II (GCPII) in a complex with CCIBzL, a urea-based inhibitor N~2~-[(1-carboxycyclopropyl)carbamoyl]-N~6~-(4-iodobenzoyl)-L-lysine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Pavlicek, J, Ptacek, J, Cerny, J, Byun, Y, Skultetyova, L, Pomper, M, Lubkowski, J, Barinka, C. | | Deposit date: | 2014-01-08 | | Release date: | 2014-05-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural characterization of P1'-diversified urea-based inhibitors of glutamate carboxypeptidase II.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4OC3
 
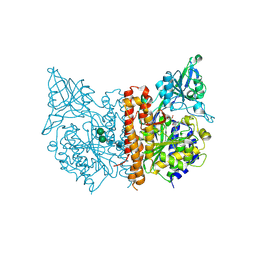 | | X-ray structure of of human glutamate carboxypeptidase II (GCPII) in a complex with CFIBzL, a urea-based inhibitor N~2~-{[(1S)-1-carboxy-2-(furan-2-yl)ethyl]carbamoyl}-N~6~-(4-iodobenzoyl)-L-lysine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pavlicek, J, Ptacek, J, Cerny, J, Byun, Y, Skultetyova, L, Pomper, M, Lubkowski, J, Barinka, C. | | Deposit date: | 2014-01-08 | | Release date: | 2014-05-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural characterization of P1'-diversified urea-based inhibitors of glutamate carboxypeptidase II.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4OC4
 
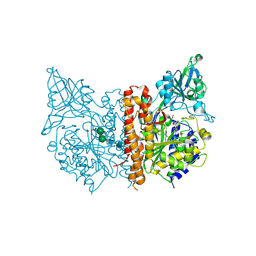 | | X-ray structure of of human glutamate carboxypeptidase II (GCPII) in a complex with CPIBzL, a urea-based inhibitor N~2~-{[(1S)-1-carboxy-2-(pyridin-4-yl)ethyl]carbamoyl}-N~6~-(4-iodobenzoyl)-L-lysine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Pavlicek, J, Ptacek, J, Cerny, J, Byun, Y, Skultetyova, L, Pomper, M, Lubkowski, J, Barinka, C. | | Deposit date: | 2014-01-08 | | Release date: | 2014-05-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural characterization of P1'-diversified urea-based inhibitors of glutamate carboxypeptidase II.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
8AQ8
 
 | | FAD-dependent monooxygenase from Stenotrophomonas maltophilia | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Maly, M, Kolenko, P, Duskova, J, Skalova, T, Dohnalek, J. | | Deposit date: | 2022-08-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Tetracycline-modifying enzyme SmTetX from Stenotrophomonas maltophilia.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
4W9Y
 
 | |
7AVC
 
 | | DoBi scaffold based on PIH1D1 N-terminal domain | | Descriptor: | GLYCEROL, PIH1 domain-containing protein 1, SODIUM ION | | Authors: | Kolenko, P, Pham, N.P, Pavlicek, J, Mikulecky, P, Schneider, B. | | Deposit date: | 2020-11-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Protein Binder (ProBi) as a New Class of Structurally Robust Non-Antibody Protein Scaffold for Directed Evolution.
Viruses, 13, 2021
|
|
2LUF
 
 | |
6T3O
 
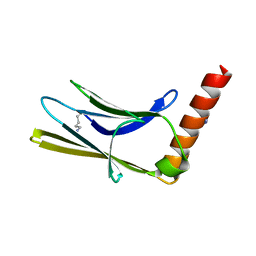 | | Crystal structure of the human myomesin domain 10 | | Descriptor: | AZIDE ION, Myomesin-1 | | Authors: | Duskova, J, Petrokova, H, Maly, P. | | Deposit date: | 2019-10-11 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Myomedin scaffold variants targeted to 10E8 HIV-1 broadly neutralizing antibody mimic gp41 epitope and elicit HIV-1 virus-neutralizing sera in mice.
Virulence, 12, 2021
|
|
2DM5
 
 | | Thermodynamic Penalty Arising From Burial of a Ligand Polar Group Within a Hydrophobic Pocket of a Protein Receptor | | Descriptor: | CADMIUM ION, Major Urinary Protein, OCTANE-1,8-DIOL | | Authors: | Barratt, E, Bronowska, A, Vondrasek, J, Bingham, R, Phillips, S, Homans, S.W. | | Deposit date: | 2006-04-20 | | Release date: | 2006-10-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thermodynamic penalty arising from burial of a ligand polar group within a hydrophobic pocket of a protein receptor
J.Mol.Biol., 362, 2006
|
|
6ROR
 
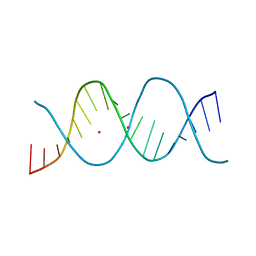 | | REP related 18-mer DNA | | Descriptor: | REP related 18-mer DNA from C. hominis, STRONTIUM ION | | Authors: | Kolenko, P, Svoboda, J, Schneider, B. | | Deposit date: | 2019-05-13 | | Release date: | 2020-07-08 | | Last modified: | 2021-01-20 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural variability of CG-rich DNA 18-mers accommodating double T-T mismatches.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6ROS
 
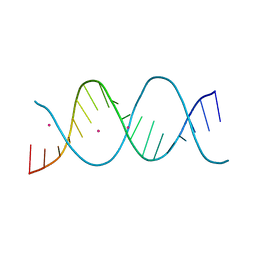 | | REP related 18-mer DNA | | Descriptor: | REP related 18-mer DNA from C. hominis, STRONTIUM ION | | Authors: | Kolenko, P, Svoboda, J, Schneider, B. | | Deposit date: | 2019-05-13 | | Release date: | 2020-07-08 | | Last modified: | 2021-01-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural variability of CG-rich DNA 18-mers accommodating double T-T mismatches.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6ROU
 
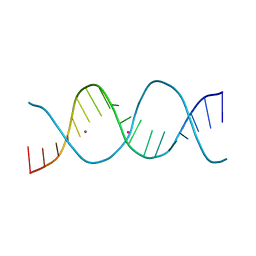 | | REP related 18-mer DNA | | Descriptor: | REP related 18-mer DNA from H. parasuis, STRONTIUM ION | | Authors: | Kolenko, P, Svoboda, J, Schneider, B. | | Deposit date: | 2019-05-13 | | Release date: | 2020-07-08 | | Last modified: | 2021-01-20 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Structural variability of CG-rich DNA 18-mers accommodating double T-T mismatches.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6SGP
 
 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) - the E424M inactive mutant, in complex with a sulfamide inhibitor GluGlu | | Descriptor: | (2~{S})-2-[[(2~{S})-1,5-bis(oxidanyl)-1,5-bis(oxidanylidene)pentan-2-yl]sulfamoylamino]pentanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Barinka, C, Shukla, S, Motlova, L. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural, Biochemical, and Computational Characterization of Sulfamides as Bimetallic Peptidase Inhibitors.
J.Chem.Inf.Model., 2024
|
|
6SKH
 
 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) - the E424M inactive mutant, in complex with a inhibitor sulfamide inhibitor GluAsp | | Descriptor: | (2~{S})-2-[[(2~{S})-1,4-bis(oxidanyl)-1,4-bis(oxidanylidene)butan-2-yl]sulfamoylamino]pentanedioic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Motlova, L, Novakova, Z, Barinka, C. | | Deposit date: | 2019-08-15 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural, Biochemical, and Computational Characterization of Sulfamides as Bimetallic Peptidase Inhibitors.
J.Chem.Inf.Model., 2024
|
|
7Z7M
 
 | | REP-related Chom18 variant with double CC mismatch | | Descriptor: | Chom18-CC DNA, STRONTIUM ION | | Authors: | Svoboda, J, Schneider, B, Berdar, D, Kolenko, P. | | Deposit date: | 2022-03-16 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformation-based refinement of 18-mer DNA structures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7Z7Z
 
 | | REP-related Chom18 variant with double TA base pair | | Descriptor: | Chom18-TA DNA, STRONTIUM ION | | Authors: | Svoboda, J, Schneider, B, Berdar, D, Kolenko, P. | | Deposit date: | 2022-03-16 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Conformation-based refinement of 18-mer DNA structures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7Z7W
 
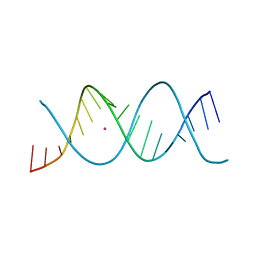 | | REP-related Chom18 variant with double GC base pairing | | Descriptor: | Chom18-GC DNA, STRONTIUM ION | | Authors: | Svoboda, J, Schneider, B, Berdar, D, Kolenko, P. | | Deposit date: | 2022-03-16 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Conformation-based refinement of 18-mer DNA structures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7Z7K
 
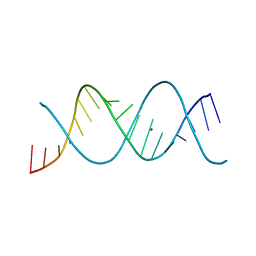 | | REP-related Chom18 variant with double AT base pairing | | Descriptor: | Chom18-AT DNA, STRONTIUM ION | | Authors: | Svoboda, J, Schneider, B, Berdar, D, Kolenko, P. | | Deposit date: | 2022-03-16 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformation-based refinement of 18-mer DNA structures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7Z7U
 
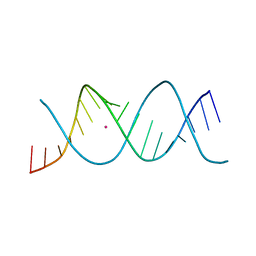 | | REP-related Chom18 variant with double CG base pair | | Descriptor: | Chom18-CG DNA, STRONTIUM ION | | Authors: | Svoboda, J, Kolenko, P, Berdar, D, Schneider, B. | | Deposit date: | 2022-03-16 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Conformation-based refinement of 18-mer DNA structures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7Z81
 
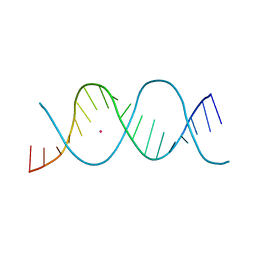 | | REP-related Chom18 variant with double TC mismatch | | Descriptor: | Chom18-TC DNA, STRONTIUM ION | | Authors: | Svoboda, J, Schneider, B, Berdar, D, Kolenko, P. | | Deposit date: | 2022-03-16 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Conformation-based refinement of 18-mer DNA structures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
