5W2J
 
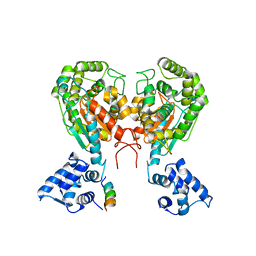 | | Crystal structure of dimeric form of mouse Glutaminase C | | Descriptor: | CHLORIDE ION, Glutaminase kidney isoform, mitochondrial, ... | | Authors: | Cerione, R.A, Li, Y. | | Deposit date: | 2017-06-06 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic Basis of Glutaminase Activation: A KEY ENZYME THAT PROMOTES GLUTAMINE METABOLISM IN CANCER CELLS.
J. Biol. Chem., 291, 2016
|
|
7SBM
 
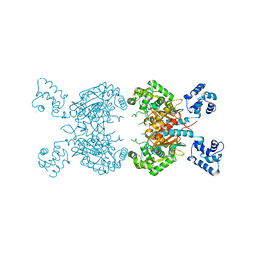 | | Human glutaminase C (Y466W) with L-Gln, open conformation | | Descriptor: | GLUTAMINE, Isoform 3 of Glutaminase kidney isoform, mitochondrial | | Authors: | Nguyen, T.-T.T, Cerione, R.A. | | Deposit date: | 2021-09-25 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | High-resolution structures of mitochondrial glutaminase C tetramers indicate conformational changes upon phosphate binding.
J.Biol.Chem., 298, 2022
|
|
7SBN
 
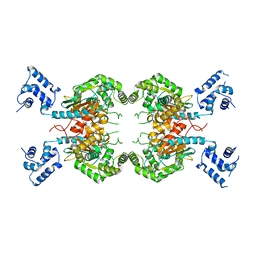 | |
7JSN
 
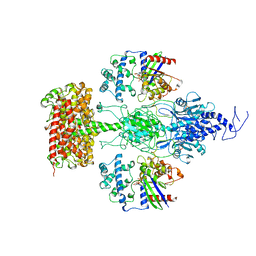 | | Structure of the Visual Signaling Complex between Transducin and Phosphodiesterase 6 | | Descriptor: | 2-{2-ETHOXY-5-[(4-ETHYLPIPERAZIN-1-YL)SULFONYL]PHENYL}-5-METHYL-7-PROPYLIMIDAZO[5,1-F][1,2,4]TRIAZIN-4(1H)-ONE, GUANOSINE-3',5'-MONOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gao, Y, Eskici, G, Ramachandran, S, Skiniotis, G, Cerione, R.A. | | Deposit date: | 2020-08-15 | | Release date: | 2020-10-21 | | Last modified: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the Visual Signaling Complex between Transducin and Phosphodiesterase 6.
Mol.Cell, 80, 2020
|
|
8SZJ
 
 | | Human glutaminase C (Y466W) with L-Gln and Pi, filamentous form | | Descriptor: | GLUTAMINE, Glutaminase kidney isoform, mitochondrial, ... | | Authors: | Feng, S, Aplin, C, Nguyen, T.-T.T, Milano, S.K, Cerione, R.A. | | Deposit date: | 2023-05-29 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Filament formation drives catalysis by glutaminase enzymes important in cancer progression.
Nat Commun, 15, 2024
|
|
8T0Z
 
 | | Human liver-type glutaminase (K253A) with L-Gln, filamentous form | | Descriptor: | GLUTAMINE, Glutaminase liver isoform, mitochondrial | | Authors: | Feng, S, Aplin, C, Nguyen, T.-T.T, Milano, S.K, Cerione, R.A. | | Deposit date: | 2023-06-01 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Filament formation drives catalysis by glutaminase enzymes important in cancer progression.
Nat Commun, 15, 2024
|
|
8SZL
 
 | | Human liver-type glutaminase (Apo form) | | Descriptor: | Glutaminase liver isoform, mitochondrial | | Authors: | Feng, S, Aplin, C, Nguyen, T.-T.T, Milano, S.K, Cerione, R.A. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Filament formation drives catalysis by glutaminase enzymes important in cancer progression.
Nat Commun, 15, 2024
|
|
8UFI
 
 | | Cryo-EM structure of bovine phosphodiesterase 6 | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, MAGNESIUM ION, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit gamma, ... | | Authors: | Aplin, C, Cerione, R.A. | | Deposit date: | 2023-10-04 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Probing the mechanism by which the retinal G protein transducin activates its biological effector PDE6.
J.Biol.Chem., 300, 2023
|
|
8ULG
 
 | | Cryo-EM structure of bovine phosphodiesterase 6 bound to IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, CYCLIC GUANOSINE MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Aplin, C, Cerione, R.A. | | Deposit date: | 2023-10-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Probing the mechanism by which the retinal G protein transducin activates its biological effector PDE6.
J.Biol.Chem., 300, 2023
|
|
8UGB
 
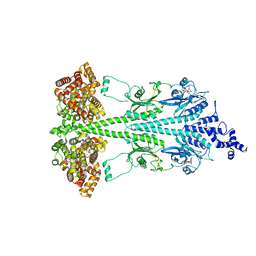 | | Cryo-EM structure of bovine phosphodiesterase 6 bound to udenafil | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, MAGNESIUM ION, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit gamma, ... | | Authors: | Aplin, C, Cerione, R.A. | | Deposit date: | 2023-10-05 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Probing the mechanism by which the retinal G protein transducin activates its biological effector PDE6.
J.Biol.Chem., 300, 2023
|
|
8UGS
 
 | | Cryo-EM structure of bovine phosphodiesterase 6 bound to cGMP | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, MAGNESIUM ION, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit gamma, ... | | Authors: | Aplin, C, Cerione, R.A. | | Deposit date: | 2023-10-06 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Probing the mechanism by which the retinal G protein transducin activates its biological effector PDE6.
J.Biol.Chem., 300, 2023
|
|
5HL1
 
 | | Crystal structure of glutaminase C in complex with inhibitor CB-839 | | Descriptor: | 2-(pyridin-2-yl)-N-(5-{4-[6-({[3-(trifluoromethoxy)phenyl]acetyl}amino)pyridazin-3-yl]butyl}-1,3,4-thiadiazol-2-yl)acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Huang, Q, Cerione, R.A. | | Deposit date: | 2016-01-14 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the clinically relevant glutaminase inhibitot CB-839 in complex with glutaminase C
To Be Published
|
|
2QRZ
 
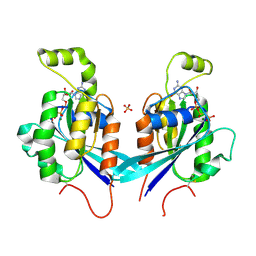 | | Cdc42 bound to GMP-PCP: Induced Fit by Effector is Required | | Descriptor: | Cell division control protein 42 homolog precursor, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Phillips, M.J, Calero, G, Chan, B, Cerione, R.A. | | Deposit date: | 2007-07-30 | | Release date: | 2008-03-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Effector Proteins Exert an Important Influence on the Signaling-active State of the Small GTPase Cdc42.
J.Biol.Chem., 283, 2008
|
|
6ULJ
 
 | | Crystal structure of human GAC in complex with inhibitor UPGL00012 | | Descriptor: | 2-phenyl-N-{6-[4-({6-[(phenylacetyl)amino]pyridazin-3-yl}oxy)piperidin-1-yl]pyridazin-3-yl}acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Huang, Q, Cerione, R.A. | | Deposit date: | 2019-10-08 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of human GAC in complex with inhibitor UPGL00045
To Be Published
|
|
6UK6
 
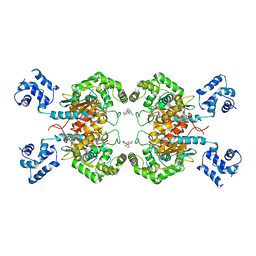 | |
6ULA
 
 | | Crystal structure of human GAC in complex with inhibitor UPGL00012 | | Descriptor: | 2-cyclopropyl-N-{5-[4-({5-[(cyclopropylacetyl)amino]-1,3,4-thiadiazol-2-yl}oxy)piperidin-1-yl]-1,3,4-thiadiazol-2-yl}acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Huang, Q, Cerione, R.A. | | Deposit date: | 2019-10-07 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of human GAC in complex with inhibitor UPGL00030
To Be Published
|
|
6UJM
 
 | | Crystal structure of human GAC in complex with inhibitor UPGL00013 | | Descriptor: | Glutaminase kidney isoform, mitochondrial, N-{5-[(3R)-3-{[5-(acetylamino)-1,3,4-thiadiazol-2-yl]amino}pyrrolidin-1-yl]-1,3,4-thiadiazol-2-yl}acetamide | | Authors: | Huang, Q, Cerione, R.A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human GAC in complex with inhibitor UPGL00013
To Be Published
|
|
6UL9
 
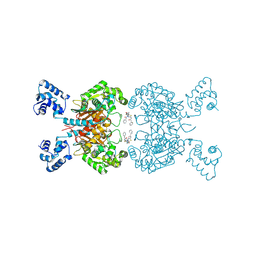 | | Crystal structure of human GAC in complex with inhibitor UPGL00023 | | Descriptor: | 2-phenyl-N-{5-[(1-{5-[(phenylacetyl)amino]-1,3,4-thiadiazol-2-yl}azetidin-3-yl)oxy]-1,3,4-thiadiazol-2-yl}acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Huang, Q.Q, Cerione, R.A. | | Deposit date: | 2019-10-07 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human GAC in complex with inhibitor UPGL00012
To Be Published
|
|
7REN
 
 | | Room temperature serial crystal structure of Glutaminase C in complex with inhibitor UPGL-00004 | | Descriptor: | 2-phenyl-N-{5-[4-({5-[(phenylacetyl)amino]-1,3,4-thiadiazol-2-yl}amino)piperidin-1-yl]-1,3,4-thiadiazol-2-yl}acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Milano, S.K, Finke, A, Cerione, R.A. | | Deposit date: | 2021-07-13 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | New insights into the molecular mechanisms of glutaminase C inhibitors in cancer cells using serial room temperature crystallography.
J.Biol.Chem., 298, 2022
|
|
7RGG
 
 | | Room temperature serial crystal structure of Glutaminase C in complex with inhibitor BPTES | | Descriptor: | Glutaminase kidney isoform, mitochondrial 68 kDa chain, N,N'-[sulfanediylbis(ethane-2,1-diyl-1,3,4-thiadiazole-5,2-diyl)]bis(2-phenylacetamide) | | Authors: | Milano, S.K, Finke, A, Cerione, R.A. | | Deposit date: | 2021-07-15 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | New insights into the molecular mechanisms of glutaminase C inhibitors in cancer cells using serial room temperature crystallography.
J.Biol.Chem., 298, 2022
|
|
6UJG
 
 | | Crystal structure of human GAC in complex with inhibitor UPGL00012 | | Descriptor: | Glutaminase kidney isoform, mitochondrial, N-{5-[(3S)-3-{[5-(acetylamino)-1,3,4-thiadiazol-2-yl]amino}pyrrolidin-1-yl]-1,3,4-thiadiazol-2-yl}acetamide | | Authors: | Huang, Q, Cerione, R.A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human GAC in complex with inhibitor UPGL00012
To Be Published
|
|
6UMD
 
 | | Crystal structure of human GAC in complex with inhibitor UPGL00012 | | Descriptor: | 2-(pyridin-3-yl)-N-(5-{4-[(5-{[(pyridin-3-yl)acetyl]amino}-1,3,4-thiadiazol-2-yl)amino]piperidin-1-yl}-1,3,4-thiadiazol-2-yl)acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Huang, Q, Cerione, R.A. | | Deposit date: | 2019-10-09 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human GAC in complex with inhibitor UPGL00012
To Be Published
|
|
6UME
 
 | | Crystal structure of human GAC in complex with inhibitor UPGL00012 | | Descriptor: | 2-(pyridin-3-yl)-N-(5-{4-[(5-{[(pyridin-3-yl)acetyl]amino}-1,3,4-thiadiazol-2-yl)amino]piperidin-1-yl}-1,3,4-thiadiazol-2-yl)acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Huang, Q, Cerione, R.A. | | Deposit date: | 2019-10-09 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of human GAC in complex with inhibitor UPGL00012
To Be Published
|
|
6UMC
 
 | | Crystal structure of human GAC in complex with inhibitor UPGL00012 | | Descriptor: | 2-phenyl-N-{5-[(3R)-3-({5-[(phenylacetyl)amino]-1,3,4-thiadiazol-2-yl}oxy)pyrrolidin-1-yl]-1,3,4-thiadiazol-2-yl}acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Huang, Q, Cerione, R.A. | | Deposit date: | 2019-10-09 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of human GAC in complex with inhibitor UPGL00012
To Be Published
|
|
6UMF
 
 | | Crystal structure of human GAC in complex with inhibitor UPGL00012 | | Descriptor: | Glutaminase kidney isoform, mitochondrial, N-(5-{[1-(5-amino-1,3,4-thiadiazol-2-yl)piperidin-4-yl]oxy}-1,3,4-thiadiazol-2-yl)-2-phenylacetamide | | Authors: | Huang, Q, Cerione, R.A. | | Deposit date: | 2019-10-09 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal structure of human GAC in complex with inhibitor UPGL00012
To Be Published
|
|
