3QWH
 
 | | Crystal structure of the 17beta-hydroxysteroid dehydrogenase from Cochliobolus lunatus in complex with NADPH and kaempferol | | Descriptor: | 17beta-hydroxysteroid dehydrogenase, 3,5,7-TRIHYDROXY-2-(4-HYDROXYPHENYL)-4H-CHROMEN-4-ONE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cassetta, A, Lamba, D, Krastanova, I, Stojan, J, Rizner, T.L, Kristan, K, Brunskole, M. | | Deposit date: | 2011-02-28 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural basis for inhibition of 17 beta-hydroxysteroid dehydrogenases by phytoestrogens: The case of fungal 17 beta-HSDcl.
J. Steroid Biochem. Mol. Biol., 171, 2017
|
|
3IS3
 
 | |
3ITD
 
 | |
3QWF
 
 | | Crystal structure of the 17beta-hydroxysteroid dehydrogenase from Cochliobolus lunatus | | Descriptor: | 17beta-hydroxysteroid dehydrogenase, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cassetta, A, Lamba, D, Krastanova, I, Stojan, J, Lanisnik-Rizner, T, Kristan, K, Brunskole, M. | | Deposit date: | 2011-02-28 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural Studies on a Fungal 17Beta-Hydroxysteroid Dehydrogenase
Biochem.J., 441, 2012
|
|
3QWI
 
 | | Crystal structure of a 17beta-hydroxysteroid dehydrogenase (holo form) from fungus Cochliobolus lunatus in complex with NADPH and coumestrol | | Descriptor: | 1,2-ETHANEDIOL, 17beta-hydroxysteroid dehydrogenase, Coumestrol, ... | | Authors: | Cassetta, A, Lamba, D, Krastanova, I, Stojan, J, Lanisnik Rizner, T, Kristan, K, Brunskole, M. | | Deposit date: | 2011-02-28 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural studies on the flavonoid inhibition of a fungal 17Beta-Hydroxysteroid dehydrogenase
Biochem.J., 441, 2012
|
|
4FJ1
 
 | | Crystal Structure of the ternary complex between a fungal 17beta-hydroxysteroid dehydrogenase (Holo form) and genistein | | Descriptor: | 17beta-hydroxysteroid dehydrogenase, DIMETHYL SULFOXIDE, GENISTEIN, ... | | Authors: | Cassetta, A, Lamba, D, Krastanova, I. | | Deposit date: | 2012-06-11 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for inhibition of 17 beta-hydroxysteroid dehydrogenases by phytoestrogens: The case of fungal 17 beta-HSDcl.
J. Steroid Biochem. Mol. Biol., 171, 2017
|
|
4FJ0
 
 | | Crystal structure of the ternary complex between a fungal 17beta-hydroxysteroid dehydrogenase (Holo form) and 3,7-dihydroxy flavone | | Descriptor: | 1,2-ETHANEDIOL, 17beta-hydroxysteroid dehydrogenase, 3,7-dihydroxy-2-phenyl-4H-chromen-4-one, ... | | Authors: | Cassetta, A, Lamba, D, Krastanova, I. | | Deposit date: | 2012-06-11 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for inhibition of 17 beta-hydroxysteroid dehydrogenases by phytoestrogens: The case of fungal 17 beta-HSDcl.
J. Steroid Biochem. Mol. Biol., 171, 2017
|
|
4FJ2
 
 | | Crystal structure of the ternary complex between a fungal 17beta-hydroxysteroid dehydrogenase (Holo form) and biochanin A | | Descriptor: | 1,2-ETHANEDIOL, 17beta-hydroxysteroid dehydrogenase, 5,7-dihydroxy-3-(4-methoxyphenyl)-4H-chromen-4-one, ... | | Authors: | Cassetta, A, Lamba, D, Krastanova, I. | | Deposit date: | 2012-06-11 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for inhibition of 17 beta-hydroxysteroid dehydrogenases by phytoestrogens: The case of fungal 17 beta-HSDcl.
J. Steroid Biochem. Mol. Biol., 171, 2017
|
|
4FIZ
 
 | |
8QIJ
 
 | | Crystallographic Structure of a Salicylate Synthase from M. abscessus (Mab-SaS) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Cassetta, A, Covaceuszach, S, Tomaiuolo, M, Meneghetti, F, Villa, S, Mori, M, Chiarelli, L.R, Mangiatordi, G.F. | | Deposit date: | 2023-09-12 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.073 Å) | | Cite: | Structural basis for specific inhibition of salicylate synthase from Mycobacterium abscessus.
Eur.J.Med.Chem., 265, 2024
|
|
5N30
 
 | | Crystal structure of the V72I mutant of the mouse alpha-Dystroglycan N-terminal region | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Dystroglycan | | Authors: | Cassetta, A, Covaceuszach, S, Brancaccio, A, Sciandra, F, Bozzi, M, Bigotti, M.G, Konarev, P.V. | | Deposit date: | 2017-02-08 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The effect of the pathological V72I, D109N and T190M missense mutations on the molecular structure of alpha-dystroglycan.
PLoS ONE, 12, 2017
|
|
5N4H
 
 | | Crystal structure of the D109N mutant of the mouse alpha-Dystroglycan N-terminal region | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Dystroglycan | | Authors: | Cassetta, A, Covaceuszach, S, Brancaccio, A, Sciandra, F, Bozzi, M, Bigotti, M.G, Konarev, P.V. | | Deposit date: | 2017-02-10 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | The effect of the pathological V72I, D109N and T190M missense mutations on the molecular structure of alpha-dystroglycan.
PLoS ONE, 12, 2017
|
|
4WIQ
 
 | | The structure of Murine alpha-Dystroglycan T190M mutant N-terminal domain. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Dystroglycan | | Authors: | Brancaccio, A, Lamba, D, Cassetta, A, Bozzi, M, Covaceuszach, S, Sciandra, F, Bigotti, M.G. | | Deposit date: | 2014-09-26 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The Structure of the T190M Mutant of Murine alpha-Dystroglycan at High Resolution: Insight into the Molecular Basis of a Primary Dystroglycanopathy.
Plos One, 10, 2015
|
|
1AH5
 
 | | REDUCED FORM SELENOMETHIONINE-LABELLED HYDROXYMETHYLBILANE SYNTHASE DETERMINED BY MAD | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, HYDROXYMETHYLBILANE SYNTHASE | | Authors: | Helliwell, J.R, Nieh, Y.P, Harrop, S.J, Cassetta, A. | | Deposit date: | 1997-04-13 | | Release date: | 1997-10-15 | | Last modified: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Determination of the structure of seleno-methionine-labelled hydroxymethylbilane synthase in its active form by multi-wavelength anomalous dispersion.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1YPN
 
 | | REDUCED FORM HYDROXYMETHYLBILANE SYNTHASE (K59Q MUTANT) CRYSTAL STRUCTURE AFTER 2 HOURS IN A FLOW CELL DETERMINED BY TIME-RESOLVED LAUE DIFFRACTION | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, HYDROXYMETHYLBILANE SYNTHASE | | Authors: | Helliwell, J.R, Nieh, Y.P, Raftery, J, Cassetta, A, Habash, J, Carr, P.D, Ursby, T, Wulff, M, Thompson, A.W, Niemann, A.C, Haedener, A. | | Deposit date: | 1998-06-26 | | Release date: | 1999-03-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Time-Resolved Structures of Hydroxymethylbilane Synthase (Lys59Gln Mutant) as It Isloaded with Substrate in the Crystal Determined by Laue Diffraction
J.Chem.Soc.,Faraday Trans., 94, 1998
|
|
1ZAN
 
 | | Crystal structure of anti-NGF AD11 Fab | | Descriptor: | CHLORIDE ION, Fab AD11 Heavy Chain, Fab AD11 Light Chain | | Authors: | Covaceuszach, S, Cattaneo, A, Cassetta, A, Lamba, D. | | Deposit date: | 2005-04-06 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Dissecting NGF interactions with TrkA and p75 receptors by structural and functional studies of an anti-NGF neutralizing antibody.
J.Mol.Biol., 381, 2008
|
|
1GTK
 
 | | Time-resolved and static-ensemble structural chemistry of hydroxymethylbilane synthase | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, PORPHOBILINOGEN DEAMINASE | | Authors: | Helliwell, J.R, Nieh, Y.P, Raftery, J, Cassetta, A, Habash, J, Carr, P.D, Ursby, T, Wulff, M, Thompson, A.W, Niemann, A.C, Haedener, A. | | Deposit date: | 2002-01-16 | | Release date: | 2003-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Time-Resolved and Static-Ensemble Structural Chemistry of Hydroxymethylbilane Synthase
Faraday Discuss., 122, 2003
|
|
5LLK
 
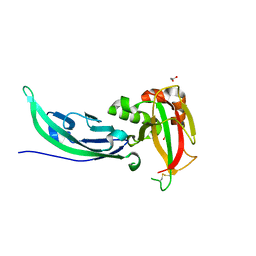 | | Crystal structure of human alpha-dystroglycan | | Descriptor: | 1,2-ETHANEDIOL, Dystroglycan | | Authors: | Covaceuszach, S, Cassetta, A, Lamba, D, Brancaccio, A, Bozzi, M, Sciandra, F, Bigotti, M.G, Konarev, P.V. | | Deposit date: | 2016-07-27 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural flexibility of human alpha-dystroglycan.
FEBS Open Bio, 7, 2017
|
|
1XT6
 
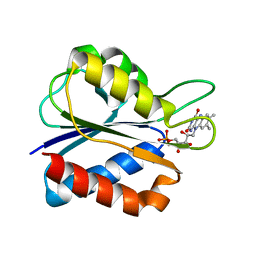 | | S35C Flavodoxin Mutant in the semiquinone state | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin | | Authors: | Artali, R, Marchini, N, Meneghetti, F, Cavazzini, D, Cassetta, A, Sassone, C, Bombieri, G, Rossi, G.L, Gilardi, G. | | Deposit date: | 2004-10-21 | | Release date: | 2004-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of S35C flavodoxin mutant from Desulfovibrio vulgaris in the semiquinone state.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3FCY
 
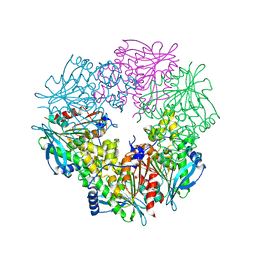 | |
3FVR
 
 | |
3FYT
 
 | |
3FVT
 
 | |
3FYU
 
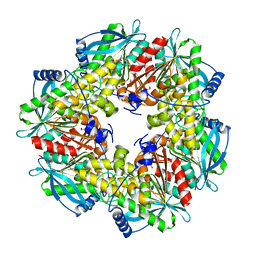 | |
7AAI
 
 | | Crystal structure of Human serum albumin in complex with perfluorooctanoic acid (PFOA) at 2.10 Angstrom Resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | Maso, L, Liberi, S, Trande, M, Angelini, A, Cendron, L. | | Deposit date: | 2020-09-04 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unveiling the binding mode of perfluorooctanoic acid to human serum albumin.
Protein Sci., 30, 2021
|
|
