3GL9
 
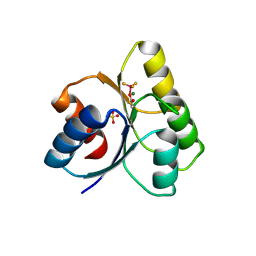 | | The structure of a histidine kinase-response regulator complex sheds light into two-component signaling and reveals a novel cis autophosphorylation mechanism | | Descriptor: | MAGNESIUM ION, Response regulator, SULFATE ION | | Authors: | Casino, P, Rubio, V, Marina, A. | | Deposit date: | 2009-03-11 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insight into Partner Specificity and Phosphoryl Transfer in Two-Component Signal Transduction
Cell(Cambridge,Mass.), 139, 2009
|
|
4KP4
 
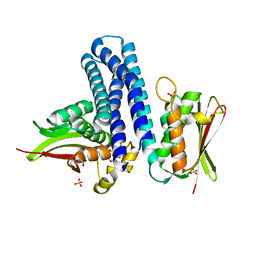 | | Deciphering cis-trans directionality and visualizing autophosphorylation in histidine kinases. | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Osmolarity sensor protein EnvZ, ... | | Authors: | Casino, P, Miguel-Romero, L, Marina, A. | | Deposit date: | 2013-05-13 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Visualizing autophosphorylation in histidine kinases.
Nat Commun, 5, 2014
|
|
6EO3
 
 | |
6EO2
 
 | |
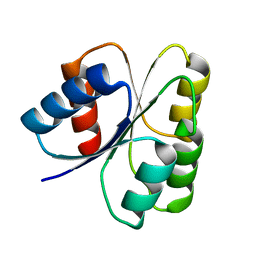
3DGF
 
 | |
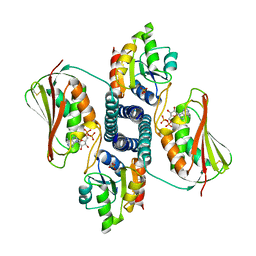
3DGE
 
 | |
5O8Z
 
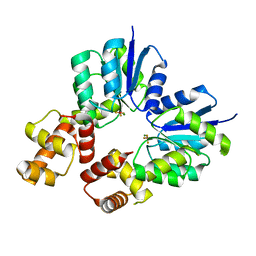 | | Conformational dynamism for DNA interaction in Salmonella typhimurium RcsB response regulator. | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Transcriptional regulatory protein RcsB | | Authors: | Casino, P, Marina, A, Miguel-Romero, L, Huesa, J. | | Deposit date: | 2017-06-14 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational dynamism for DNA interaction in the Salmonella RcsB response regulator.
Nucleic Acids Res., 46, 2018
|
|
5O8Y
 
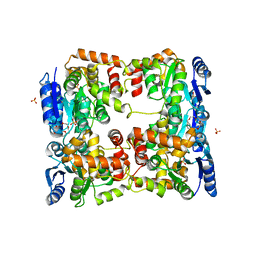 | | Conformational dynamism for DNA interaction in Salmonella typhimurium RcsB response regulator. | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, SULFATE ION, Transcriptional regulatory protein RcsB | | Authors: | Casino, P, Marina, A, Miguel-Romero, L, Huesa, J. | | Deposit date: | 2017-06-14 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational dynamism for DNA interaction in the Salmonella RcsB response regulator.
Nucleic Acids Res., 46, 2018
|
|
5NAY
 
 | |
5NAX
 
 | |
5NB2
 
 | |
5NAZ
 
 | |
5NB0
 
 | |
5NB1
 
 | |
6Z0J
 
 | |
6Z0K
 
 | | Crystal structure of laccase from Pediococcus acidilactici Pp5930 (Hepes pH 7.5) | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, Putative multicopper oxidase mco | | Authors: | Casino, P, Huesa, J, Pardo, I. | | Deposit date: | 2020-05-09 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis and biochemical properties of laccase enzymes from two Pediococcus species.
Microb Biotechnol, 14, 2021
|
|
4JAU
 
 | | Structural basis of a rationally rewired protein-protein interface (HK853mutant A268V, A271G, T275M, V294T and D297E) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Histidine kinase | | Authors: | Podgornaia, A.I, Casino, P, Marina, A, Laub, M.T. | | Deposit date: | 2013-02-19 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of a rationally rewired protein-protein interface critical to bacterial signaling
Structure, 21, 2013
|
|
4JAS
 
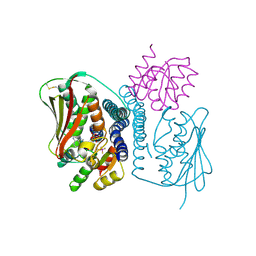 | | Structural basis of a rationally rewired protein-protein interface (HK853mutant A268V, A271G, T275M, V294T and D297E and RR468mutant V13P, L14I, I17M and N21V) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Histidine kinase, MAGNESIUM ION, ... | | Authors: | Podgornaia, A.I, Casino, P, Marina, A, Laub, M.T. | | Deposit date: | 2013-02-19 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of a rationally rewired protein-protein interface critical to bacterial signaling
Structure, 21, 2013
|
|
4JA2
 
 | | Structural basis of a rationally rewired protein-protein interface (RR468mutant V13P, L14I, I17M and N21V) | | Descriptor: | ACETATE ION, MAGNESIUM ION, Response regulator, ... | | Authors: | Podgornaia, A.I, Casino, P, Marina, A, Laub, M.T. | | Deposit date: | 2013-02-18 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural basis of a rationally rewired protein-protein interface critical to bacterial signaling
Structure, 21, 2013
|
|
4JAV
 
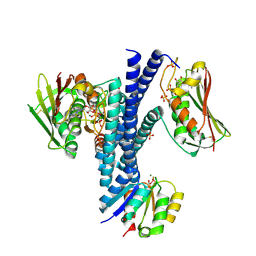 | | Structural basis of a rationally rewired protein-protein interface (HK853wt and RR468mutant V13P, L14I, I17M and N21V) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Histidine kinase, ... | | Authors: | Podgornaia, A.I, Casino, P, Marina, A, Laub, M.T. | | Deposit date: | 2013-02-19 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of a rationally rewired protein-protein interface critical to bacterial signaling
Structure, 21, 2013
|
|
6ZIX
 
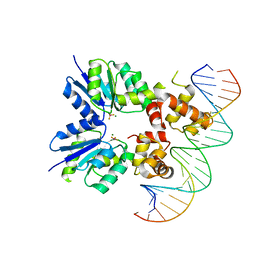 | | Structure of RcsB from Salmonella enterica serovar Typhimurium bound to promoter P1flhDC in the presence of phosphomimetic BeF3- | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, P1flhDC promoter sequence of 23 bp, ... | | Authors: | Huesa, J, Marina, A, Casino, P. | | Deposit date: | 2020-06-26 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure-based analyses of Salmonella RcsB variants unravel new features of the Rcs regulon.
Nucleic Acids Res., 49, 2021
|
|
6ZIL
 
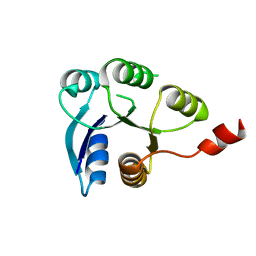 | |
6ZJ2
 
 | | Structure of RcsB from Salmonella enterica serovar Typhimurium bound to promoter rprA in the presence of phosphomimetic BeF3- | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, DNA (5'-D(P*CP*CP*GP*AP*TP*CP*AP*GP*AP*TP*TP*CP*GP*TP*CP*TP*CP*AP*AP*TP*AP*GP*G)-3'), MAGNESIUM ION, ... | | Authors: | Huesa, J, Marina, A, Casino, P. | | Deposit date: | 2020-06-27 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Structure-based analyses of Salmonella RcsB variants unravel new features of the Rcs regulon.
Nucleic Acids Res., 49, 2021
|
|
6ZII
 
 | |
5LWK
 
 | | MaeR response regulator bound to beryllium trifluoride | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Transcriptional regulatory protein | | Authors: | Miguel-Romero, L, Casino, P, Landete, J.M, Monedero, V, Zuniga, M, Marina, A. | | Deposit date: | 2016-09-18 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The malate sensing two-component system MaeKR is a non-canonical class of sensory complex for C4-dicarboxylates.
Sci Rep, 7, 2017
|
|
