6EM9
 
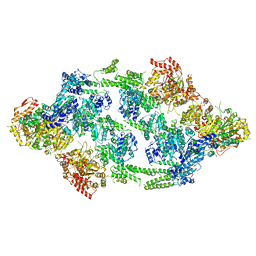 | | S.aureus ClpC resting state, asymmetric map | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpC | | Authors: | Carroni, M, Mogk, A, Bukau, B, Franke, K. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Last modified: | 2018-11-28 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Regulatory coiled-coil domains promote head-to-head assemblies of AAA+ chaperones essential for tunable activity control.
Elife, 6, 2017
|
|
6EMW
 
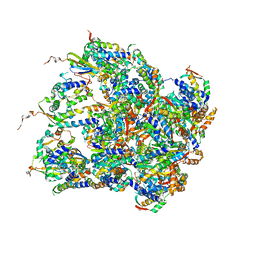 | | Structure of S.aureus ClpC in complex with MecA | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit, ATP-dependent Clp protease ATP-binding subunit ClpC, Adapter protein MecA, ... | | Authors: | Carroni, M, Mogk, A, Bukau, B, Franke, K. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-27 | | Last modified: | 2019-09-25 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Regulatory coiled-coil domains promote head-to-head assemblies of AAA+ chaperones essential for tunable activity control.
Elife, 6, 2017
|
|
6EM8
 
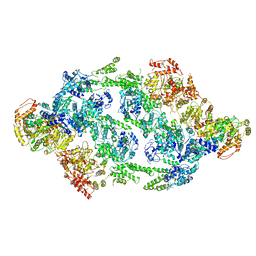 | | S.aureus ClpC resting state, C2 symmetrised | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpC | | Authors: | Carroni, M, Mogk, A. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Regulatory coiled-coil domains promote head-to-head assemblies of AAA+ chaperones essential for tunable activity control.
Elife, 6, 2017
|
|
4D2U
 
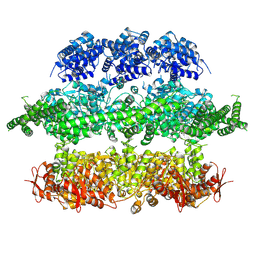 | | Negative-stain electron microscopy of E. coli ClpB (BAP form bound to ClpP) | | Descriptor: | CHAPERONE PROTEIN CLPB | | Authors: | Carroni, M, Kummer, E, Oguchi, Y, Clare, D.K, Wendler, P, Sinning, I, Kopp, J, Mogk, A, Bukau, B, Saibil, H.R. | | Deposit date: | 2014-05-13 | | Release date: | 2014-06-04 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Head-to-Tail Interactions of the Coiled-Coil Domains Regulate Clpb Activity and Cooperation with Hsp70 in Protein Disaggregation.
Elife, 3, 2014
|
|
4D2Q
 
 | | Negative-stain electron microscopy of E. coli ClpB mutant E432A (BAP form bound to ClpP) | | Descriptor: | CLPB | | Authors: | Carroni, M, Kummer, E, Oguchi, Y, Clare, D.K, Wendler, P, Sinning, I, Kopp, J, Mogk, A, Bukau, B, Saibil, H.R. | | Deposit date: | 2014-05-12 | | Release date: | 2014-06-04 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Head-to-Tail Interactions of the Coiled-Coil Domains Regulate Clpb Activity and Cooperation with Hsp70 in Protein Disaggregation.
Elife, 3, 2014
|
|
4D2X
 
 | | Negative-stain electron microscopy of E. coli ClpB of Y503D hyperactive mutant (BAP form bound to ClpP) | | Descriptor: | CHAPERONE PROTEIN CLPB | | Authors: | Carroni, M, Kummer, E, Oguchi, Y, Clare, D.K, Wendler, P, Sinning, I, Kopp, J, Mogk, A, Bukau, B, Saibil, H.R. | | Deposit date: | 2014-05-13 | | Release date: | 2014-06-04 | | Last modified: | 2019-01-23 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Head-to-Tail Interactions of the Coiled-Coil Domains Regulate Clpb Activity and Cooperation with Hsp70 in Protein Disaggregation.
Elife, 3, 2014
|
|
7NTM
 
 | |
6TQL
 
 | | Cryo-EM of elastase-treated human uromodulin (UMOD)/Tamm-Horsfall protein (THP) filament | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Stsiapanava, A, Xu, C, Carroni, M, Wu, B, Jovine, L. | | Deposit date: | 2019-12-16 | | Release date: | 2020-11-04 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Cryo-EM structure of native human uromodulin, a zona pellucida module polymer.
Embo J., 39, 2020
|
|
5OFO
 
 | | Cryo EM structure of the E. coli disaggregase ClpB (BAP form, DWB mutant), in the ATPgammaS state, bound to the model substrate casein | | Descriptor: | Chaperone protein ClpB,ATP-dependent Clp protease ATP-binding subunit ClpA,Chaperone protein ClpB, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Deville, C, Carroni, M, Franke, K.B, Topf, M, Bukau, B, Mogk, A, Saibil, H.R. | | Deposit date: | 2017-07-11 | | Release date: | 2017-08-16 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural pathway of regulated substrate transfer and threading through an Hsp100 disaggregase.
Sci Adv, 3, 2017
|
|
5OG1
 
 | | Cryo EM structure of the E. coli disaggregase ClpB (BAP form, DWB mutant), in the ATPgammaS state | | Descriptor: | Chaperone protein ClpB,ATP-dependent Clp protease ATP-binding subunit ClpA,Chaperone protein ClpB, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Deville, C, Carroni, M, Franke, K.B, Topf, M, Bukau, B, Mogk, A, Saibil, H.R. | | Deposit date: | 2017-07-11 | | Release date: | 2017-08-16 | | Last modified: | 2019-08-21 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural pathway of regulated substrate transfer and threading through an Hsp100 disaggregase.
Sci Adv, 3, 2017
|
|
6TQK
 
 | | Cryo-EM of native human uromodulin (UMOD)/Tamm-Horsfall protein (THP) filament. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, ... | | Authors: | Stsiapanava, A, Xu, C, Carroni, M, Wu, B, Jovine, L. | | Deposit date: | 2019-12-16 | | Release date: | 2020-11-04 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cryo-EM structure of native human uromodulin, a zona pellucida module polymer.
Embo J., 39, 2020
|
|
7PFP
 
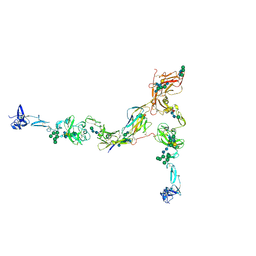 | | Full-length cryo-EM structure of the native human uromodulin (UMOD)/Tamm-Horsfall protein (THP) filament | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jovine, L, Xu, C, Stsiapanava, A, Carroni, M, Tunyasuvunakool, K, Jumper, J, Wu, B. | | Deposit date: | 2021-08-11 | | Release date: | 2022-03-16 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7Q3N
 
 | | Cryo-EM of the complex between human uromodulin (UMOD)/Tamm-Horsfall protein (THP) and the FimH lectin domain from uropathogenic E. coli | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Type 1 fimbiral adhesin FimH, ... | | Authors: | Jovine, L, Xu, C, Stsiapanava, A, Carroni, M, Tunyasuvunakool, K, Jumper, J, Wu, B. | | Deposit date: | 2021-10-28 | | Release date: | 2022-03-16 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6ZGK
 
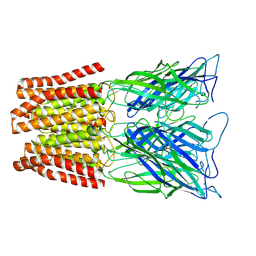 | | GLIC pentameric ligand-gated ion channel, pH 3 | | Descriptor: | Proton-gated ion channel | | Authors: | Rovsnik, U, Zhuang, Y, Forsberg, B.O, Carroni, M, Yvonnesdotter, L, Howard, R.J, Lindahl, E. | | Deposit date: | 2020-06-18 | | Release date: | 2021-05-26 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Dynamic closed states of a ligand-gated ion channel captured by cryo-EM and simulations.
Life Sci Alliance, 4, 2021
|
|
6ZGD
 
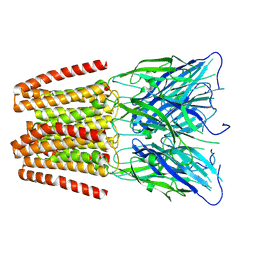 | | GLIC pentameric ligand-gated ion channel, pH 7 | | Descriptor: | Proton-gated ion channel | | Authors: | Rovsnik, U, Zhuang, Y, Forsberg, B.O, Carroni, M, Yvonnesdotter, L, Howard, R.J, Lindahl, E. | | Deposit date: | 2020-06-18 | | Release date: | 2021-05-26 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Dynamic closed states of a ligand-gated ion channel captured by cryo-EM and simulations.
Life Sci Alliance, 4, 2021
|
|
6ZGJ
 
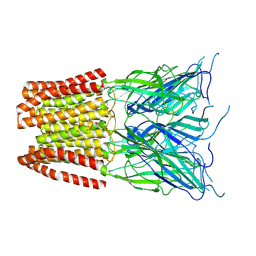 | | GLIC pentameric ligand-gated ion channel, pH 5 | | Descriptor: | Proton-gated ion channel | | Authors: | Rovsnik, U, Zhuang, Y, Forsberg, B.O, Carroni, M, Yvonnesdotter, L, Howard, R.J, Lindahl, E. | | Deposit date: | 2020-06-18 | | Release date: | 2021-05-26 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Dynamic closed states of a ligand-gated ion channel captured by cryo-EM and simulations.
Life Sci Alliance, 4, 2021
|
|
6T8H
 
 | | Cryo-EM structure of the DNA-bound PolD-PCNA processive complex from P. abyssi | | Descriptor: | DNA polymerase II small subunit, DNA polymerase sliding clamp, DNA primer, ... | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Pehau-Arnaudet, G, England, P, Lindhal, E, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-24 | | Release date: | 2020-03-04 | | Last modified: | 2020-04-08 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
6T7Y
 
 | | Structure of PCNA bound to cPIP motif of DP2 from P. abyssi | | Descriptor: | DNA polymerase sliding clamp, cPIP motif from the DP2 large subunit of PolD | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
6T7X
 
 | | Crystal structure of PCNA from P. abyssi | | Descriptor: | DNA polymerase sliding clamp | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
6HMS
 
 | | Cryo-EM map of DNA polymerase D from Pyrococcus abyssi in complex with DNA | | Descriptor: | DNA (5'-D(*GP*AP*GP*AP*CP*GP*GP*GP*CP*CP*GP*CP*GP*TP*C)-3'), DNA (5'-D(P*TP*GP*AP*CP*GP*CP*GP*GP*CP*CP*CP*GP*TP*CP*TP*C)-3'), DNA polymerase II large subunit,DNA polymerase II large subunit, ... | | Authors: | Raia, P, Carroni, M, Sauguet, L. | | Deposit date: | 2018-09-12 | | Release date: | 2019-01-30 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Structure of the DP1-DP2 PolD complex bound with DNA and its implications for the evolutionary history of DNA and RNA polymerases.
PLoS Biol., 17, 2019
|
|
7PGP
 
 | |
7PGQ
 
 | | GAP-SecPH region of human neurofibromin isoform 2 in closed conformation. | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, Neurofibromin, ZINC ION | | Authors: | Naschberger, A, Baradaran, R, Carroni, M, Rupp, B. | | Deposit date: | 2021-08-15 | | Release date: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The structure of neurofibromin isoform 2 reveals different functional states.
Nature, 599, 2021
|
|
6FU8
 
 | |
7PGT
 
 | | The structure of human neurofibromin isoform 2 in opened conformation. | | Descriptor: | Neurofibromin, ZINC ION | | Authors: | Naschberger, A, Baradaran, R, Carroni, M, Rupp, B. | | Deposit date: | 2021-08-15 | | Release date: | 2021-11-17 | | Last modified: | 2021-11-24 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | The structure of neurofibromin isoform 2 reveals different functional states.
Nature, 599, 2021
|
|
7PGU
 
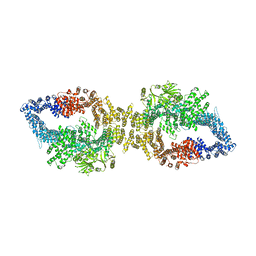 | | Autoinhibited structure of human neurofibromin isoform 2 stabilized by Zinc. | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, Neurofibromin, ZINC ION | | Authors: | Naschberger, A, Baradaran, R, Carroni, M, Rupp, B. | | Deposit date: | 2021-08-15 | | Release date: | 2021-11-17 | | Last modified: | 2021-11-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structure of neurofibromin isoform 2 reveals different functional states.
Nature, 599, 2021
|
|
