2BO5
 
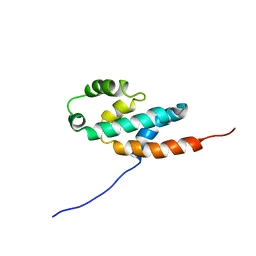 | | Bovine oligomycin sensitivity conferral protein N-terminal domain | | Descriptor: | ATP SYNTHASE OLIGOMYCIN SENSITIVITY CONFERRAL PROTEIN | | Authors: | Carbajo, R.J, Kellas, F.A, Runswick, M.J, Montgomery, M.G, Walker, J.E, Neuhaus, D. | | Deposit date: | 2005-04-07 | | Release date: | 2005-08-17 | | Last modified: | 2018-05-09 | | Method: | SOLUTION NMR | | Cite: | Structure of the F1-binding domain of the stator of bovine F1Fo-ATPase and how it binds an alpha-subunit.
J. Mol. Biol., 351, 2005
|
|
1VZS
 
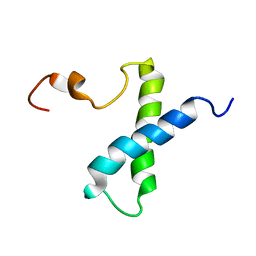 | | Solution structure of subunit F6 from the peripheral stalk region of ATP synthase from bovine heart mitochondria | | Descriptor: | ATP SYNTHASE COUPLING FACTOR 6, MITOCHONDRIAL PRECURSOR | | Authors: | Carbajo, R.J, Silvester, J.A, Runswick, M.J, Walker, J.E, Neuhaus, D. | | Deposit date: | 2004-05-25 | | Release date: | 2004-09-02 | | Last modified: | 2018-01-24 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Subunit F(6) from the Peripheral Stalk Region of ATP Synthase from Bovine Heart Mitochondria
J.Mol.Biol., 342, 2004
|
|
2W9V
 
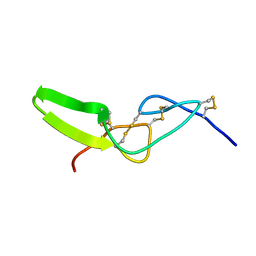 | | Solution structure of jerdostatin from Trimeresurus jerdonii with end C-terminal residues N45G46 deleted | | Descriptor: | SHORT DISINTEGRIN JERDOSTATIN | | Authors: | Carbajo, R.J, Sanz, L, Mosulen, S, Calvete, J.J, Pineda-Lucena, A. | | Deposit date: | 2009-01-29 | | Release date: | 2010-03-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR Structure and Dynamics of Recombinant Wild-Type and Mutated Jerdostatin, a Selective Inhibitor of Integrin Alpha1 Beta1
Proteins, 79, 2011
|
|
2W9U
 
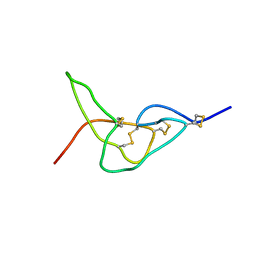 | | Solution structure of jerdostatin mutant R24K from Trimeresurus jerdonii | | Descriptor: | SHORT DISINTEGRIN JERDOSTATIN | | Authors: | Carbajo, R.J, Sanz, L, Mosulen, S, Calvete, J.J, Pineda-Lucena, A. | | Deposit date: | 2009-01-29 | | Release date: | 2010-03-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR Structure and Dynamics of Recombinant Wild-Type and Mutated Jerdostatin, a Selective Inhibitor of Integrin Alpha1 Beta1
Proteins, 79, 2011
|
|
2W9O
 
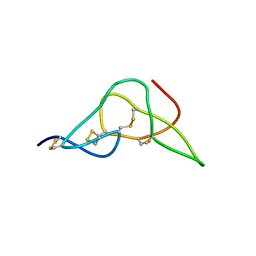 | | Solution structure of jerdostatin from Trimeresurus jerdonii | | Descriptor: | SHORT DISINTEGRIN JERDOSTATIN | | Authors: | Carbajo, R.J, Sanz, L, Mosulen, S, Calvete, J.J, Pineda-Lucena, A. | | Deposit date: | 2009-01-27 | | Release date: | 2010-03-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR Structure and Dynamics of Recombinant Wild-Type and Mutated Jerdostatin, a Selective Inhibitor of Integrin Alpha1 Beta1
Proteins, 79, 2011
|
|
2W9W
 
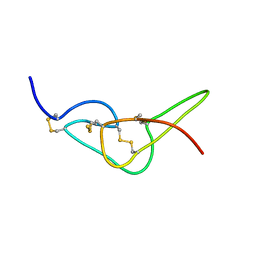 | | Solution structure of jerdostatin mutant R24K from Trimeresurus jerdonii with end C-terminal residues N45G46 deleted | | Descriptor: | SHORT DISINTEGRIN JERDOSTATIN | | Authors: | Carbajo, R.J, Sanz, L, Mosulen, S, Calvete, J.J, Pineda-Lucena, A. | | Deposit date: | 2009-01-29 | | Release date: | 2010-03-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR Structure and Dynamics of Recombinant Wild-Type and Mutated Jerdostatin, a Selective Inhibitor of Integrin Alpha1 Beta1
Proteins, 79, 2011
|
|
2JMX
 
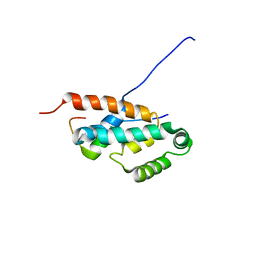 | | OSCP-NT (1-120) in complex with N-terminal (1-25) alpha subunit from F1-ATPase | | Descriptor: | ATP synthase O subunit, mitochondrial, ATP synthase subunit alpha heart isoform | | Authors: | Carbajo, R.J, Neuhaus, D, Kellas, F.A, Yang, J, Runswick, M.J, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2006-12-12 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | How the N-terminal Domain of the OSCP Subunit of Bovine F(1)F(o)-ATP Synthase Interacts with the N-terminal Region of an Alpha Subunit
J.Mol.Biol., 368, 2007
|
|
2MP5
 
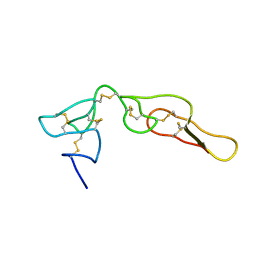 | | Structure of Bitistatin B | | Descriptor: | Disintegrin bitistatin | | Authors: | Carbajo, R.J, Calvete, J, Sanz, L, Perez, A. | | Deposit date: | 2014-05-11 | | Release date: | 2014-11-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structure of bitistatin - a missing piece in the evolutionary pathway of snake venom disintegrins.
Febs J., 282, 2015
|
|
2MOP
 
 | | Structure of Bitistatin A | | Descriptor: | Disintegrin bitistatin | | Authors: | Carbajo, R.J, Calvete, J, Sanz, L, Perez, A. | | Deposit date: | 2014-04-29 | | Release date: | 2014-11-12 | | Last modified: | 2015-02-25 | | Method: | SOLUTION NMR | | Cite: | NMR structure of bitistatin - a missing piece in the evolutionary pathway of snake venom disintegrins.
Febs J., 282, 2015
|
|
2J10
 
 | | p53 tetramerization domain mutant T329F Q331K | | Descriptor: | CELLULAR TUMOR ANTIGEN P53 | | Authors: | Carbajo, R.J, Mora, P, Sanchez del Pino, M.M, Perez-Paya, E, Pineda-Lucena, A. | | Deposit date: | 2006-08-08 | | Release date: | 2007-08-28 | | Last modified: | 2018-04-25 | | Method: | SOLUTION NMR | | Cite: | Solvent-exposed residues located in the beta-sheet modulate the stability of the tetramerization domain of p53--a structural and combinatorial approach.
Proteins, 71, 2008
|
|
2J11
 
 | | p53 tetramerization domain mutant Y327S T329G Q331G | | Descriptor: | CELLULAR TUMOR ANTIGEN P53 | | Authors: | Carbajo, R.J, Mora, P, Sanchez del Pino, M.M, Perez-Paya, E, Pineda-Lucena, A. | | Deposit date: | 2006-08-08 | | Release date: | 2007-08-28 | | Last modified: | 2018-04-25 | | Method: | SOLUTION NMR | | Cite: | Solvent-exposed residues located in the beta-sheet modulate the stability of the tetramerization domain of p53--a structural and combinatorial approach.
Proteins, 71, 2008
|
|
2J0Z
 
 | | p53 tetramerization domain wild type | | Descriptor: | CELLULAR TUMOR ANTIGEN P53 | | Authors: | Carbajo, R.J, Mora, P, Sanchez del Pino, M.M, Perez-Paya, E, Pineda-Lucena, A. | | Deposit date: | 2006-08-08 | | Release date: | 2007-08-28 | | Last modified: | 2018-04-25 | | Method: | SOLUTION NMR | | Cite: | Solvent-exposed residues located in the beta-sheet modulate the stability of the tetramerization domain of p53--a structural and combinatorial approach.
Proteins, 71, 2008
|
|
6IAR
 
 | | Tricyclic indazoles a novel class of selective estrogen receptor degrader antagonists | | Descriptor: | 3-[4-[(6~{R})-7-(2-methylpropyl)-3,6,8,9-tetrahydropyrazolo[4,3-f]isoquinolin-6-yl]phenyl]propanoic acid, Estrogen receptor | | Authors: | Scott, J.S, Bailey, A, Buttar, D, Carbajo, R.J, Curwen, J, Davies, R.D.M, Degorce, S.L, Donald, C, Gangl, E, Greenwood, R, Groombridge, S.D, Johnson, T, Lamont, S, Lawson, M, Lister, A, Morrow, C, Moss, T, Pink, J.H, Polanski, R. | | Deposit date: | 2018-11-27 | | Release date: | 2019-01-23 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Tricyclic Indazoles-A Novel Class of Selective Estrogen Receptor Degrader Antagonists.
J.Med.Chem., 62, 2019
|
|
1HF9
 
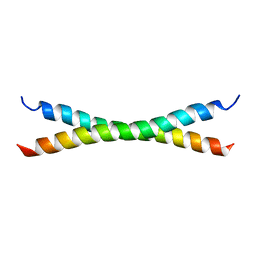 | | C-Terminal Coiled-Coil Domain from Bovine IF1 | | Descriptor: | ATPASE INHIBITOR (MITOCHONDRIAL) | | Authors: | Gordon-Smith, D.J, Carbajo, R.J, Yang, J.-C, Videler, H, Runswick, M.J, Walker, J.E, Neuhaus, D. | | Deposit date: | 2000-11-30 | | Release date: | 2001-05-31 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a C-terminal coiled-coil domain from bovine IF(1): the inhibitor protein of F(1) ATPase.
J. Mol. Biol., 308, 2001
|
|
8FU3
 
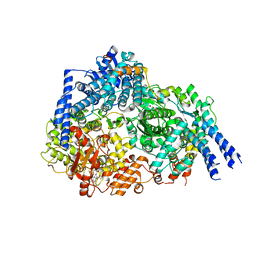 | | Structure Of Respiratory Syncytial Virus Polymerase with Novel Non-Nucleoside Inhibitor | | Descriptor: | 8-methoxy-3-methyl-N-{(2S)-3,3,3-trifluoro-2-[5-fluoro-6-(4-fluorophenyl)-4-(2-hydroxypropan-2-yl)pyridin-2-yl]-2-hydroxypropyl}cinnoline-6-carboxamide, Phosphoprotein, RNA-directed RNA polymerase L | | Authors: | Yu, X, Abeywickrema, P, Bonneux, B, Behera, I, Jacoby, E, Fung, A, Adhikary, S, Bhaumik, A, Carbajo, R.J, Bruyn, S.D, Miller, R, Patrick, A, Pham, Q, Piassek, M, Verheyen, N, Shareef, A, Sutto-Ortiz, P, Ysebaert, N, Vlijmen, H.V, Jonckers, T.H.M, Herschke, F, McLellan, J.S, Decroly, E, Fearns, R, Grosse, S, Roymans, D, Sharma, S, Rigaux, P, Jin, Z. | | Deposit date: | 2023-01-16 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural and mechanistic insights into the inhibition of respiratory syncytial virus polymerase by a non-nucleoside inhibitor.
Commun Biol, 6, 2023
|
|
6RFJ
 
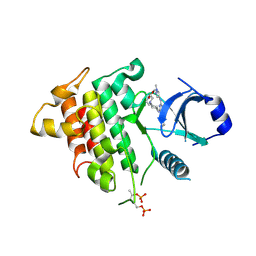 | | IRAK4 IN COMPLEX WITH inhibitor | | Descriptor: | Interleukin-1 receptor-associated kinase 4, SULFATE ION, methyl 4-[4-[[6-(cyanomethyl)-2-[(1-methylpyrazol-4-yl)amino]pyrido[3,2-d]pyrimidin-4-yl]amino]cyclohexyl]piperazine-1-carboxylate | | Authors: | Xue, Y, Degorce, S.L, Robb, G.R, Ferguson, A.D. | | Deposit date: | 2019-04-15 | | Release date: | 2019-10-30 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Discovery of a Series of 5-Azaquinazolines as Orally Efficacious IRAK4 Inhibitors Targeting MyD88L265PMutant Diffuse Large B Cell Lymphoma.
J.Med.Chem., 62, 2019
|
|
6RFI
 
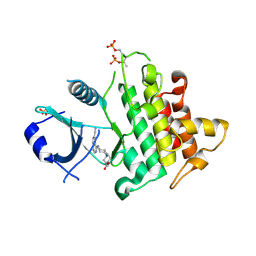 | | IRAK4 IN COMPLEX WITH inhibitor | | Descriptor: | Interleukin-1 receptor-associated kinase 4, SULFATE ION, methyl 4-[4-[(6-cyanoquinazolin-4-yl)amino]cyclohexyl]piperazine-1-carboxylate | | Authors: | Xue, Y, Degorce, S.L, Robb, G.R, Ferguson, A.D. | | Deposit date: | 2019-04-15 | | Release date: | 2019-10-30 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Discovery of a Series of 5-Azaquinazolines as Orally Efficacious IRAK4 Inhibitors Targeting MyD88L265PMutant Diffuse Large B Cell Lymphoma.
J.Med.Chem., 62, 2019
|
|
6SQ0
 
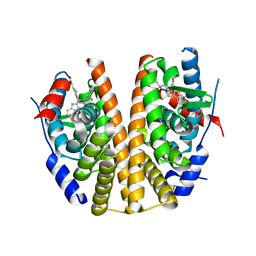 | |
6SUO
 
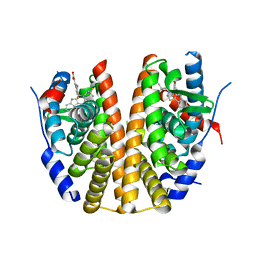 | | ERa_L536S (L536S/C381S/C471S,C530S) in complex with a tricyclic indole (compound 6) | | Descriptor: | (~{E})-3-[3,5-bis(fluoranyl)-4-[(1~{R},3~{R})-2-(2-fluoranyl-2-methyl-propyl)-1,3-dimethyl-4,9-dihydro-3~{H}-pyrido[3,4-b]indol-1-yl]phenyl]prop-2-enoic acid, Estrogen receptor | | Authors: | Breed, J. | | Deposit date: | 2019-09-16 | | Release date: | 2019-10-30 | | Last modified: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Building Bridges in a Series of Estrogen Receptor Degraders: An Application of Metathesis in Medicinal Chemistry.
Acs Med.Chem.Lett., 10, 2019
|
|
6T5B
 
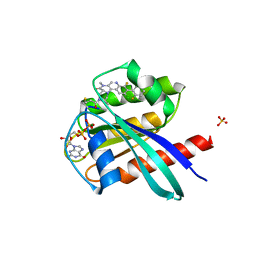 | | KRasG12C ligand complex | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Phillips, C. | | Deposit date: | 2019-10-15 | | Release date: | 2020-02-26 | | Last modified: | 2020-05-27 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structure-Based Design and Pharmacokinetic Optimization of Covalent Allosteric Inhibitors of the Mutant GTPase KRASG12C.
J.Med.Chem., 63, 2020
|
|
6T5U
 
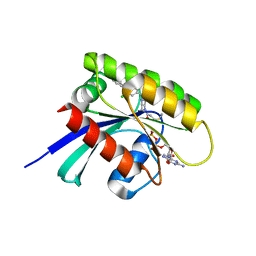 | | KRasG12C ligand complex | | Descriptor: | 1-[(7R)-16-chloro-15-(5-methyl-1H-indazol-4-yl)-9-oxa-2,5,12-triazatetracyclo[8.8.0.02,7.013,18]octadeca-1(10),11,13,15,17-pentaen-5-yl]prop-2-en-1-one, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Phillips, C. | | Deposit date: | 2019-10-17 | | Release date: | 2020-02-19 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure-Based Design and Pharmacokinetic Optimization of Covalent Allosteric Inhibitors of the Mutant GTPase KRASG12C.
J.Med.Chem., 63, 2020
|
|
6T5V
 
 | | KRasG12C ligand complex | | Descriptor: | 1-[4-[6-chloranyl-7-(5-methyl-1~{H}-indazol-4-yl)quinazolin-4-yl]piperazin-1-yl]propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Phillips, C. | | Deposit date: | 2019-10-17 | | Release date: | 2020-02-19 | | Last modified: | 2020-05-27 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structure-Based Design and Pharmacokinetic Optimization of Covalent Allosteric Inhibitors of the Mutant GTPase KRASG12C.
J.Med.Chem., 63, 2020
|
|
5N21
 
 | | Crystal structure of the BCL6 BTB domain in complex with pyrazolo-pyrimidine ligand | | Descriptor: | 2-[(2~{S})-1-[3-cyano-7-[(2-oxidanylidene-3,4-dihydro-1~{H}-quinolin-6-yl)amino]pyrazolo[1,5-a]pyrimidin-5-yl]pyrrolidin-2-yl]ethanoic acid, B-cell lymphoma 6 protein, CHLORIDE ION | | Authors: | Robb, G, Ferguson, A, Hargreaves, D. | | Deposit date: | 2017-02-06 | | Release date: | 2017-05-17 | | Last modified: | 2017-06-07 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Discovery of Pyrazolo[1,5-a]pyrimidine B-Cell Lymphoma 6 (BCL6) Binders and Optimization to High Affinity Macrocyclic Inhibitors.
J. Med. Chem., 60, 2017
|
|
5N1Z
 
 | | Crystal structure of the BCL6 BTB domain in complex with pyrazolo-pyrimidine macrocyclic ligand | | Descriptor: | B-cell lymphoma 6 protein, CHLORIDE ION, pyrazolo-pyrimidine macrocycle | | Authors: | Robb, G, Ferguson, A, Hargreaves, D. | | Deposit date: | 2017-02-06 | | Release date: | 2017-05-17 | | Last modified: | 2017-06-07 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery of Pyrazolo[1,5-a]pyrimidine B-Cell Lymphoma 6 (BCL6) Binders and Optimization to High Affinity Macrocyclic Inhibitors.
J. Med. Chem., 60, 2017
|
|
5N1X
 
 | | Crystal structure of the BCL6 BTB domain in complex with pyrazolo-pyrimidine ligand | | Descriptor: | B-cell lymphoma 6 protein, ~{N}-ethyl-5-pyridin-3-yl-pyrazolo[1,5-a]pyrimidin-7-amine | | Authors: | Robb, G, Ferguson, A, Hargreaves, D. | | Deposit date: | 2017-02-06 | | Release date: | 2017-05-17 | | Last modified: | 2017-06-07 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of Pyrazolo[1,5-a]pyrimidine B-Cell Lymphoma 6 (BCL6) Binders and Optimization to High Affinity Macrocyclic Inhibitors.
J. Med. Chem., 60, 2017
|
|
