6VW2
 
 | | Cryo-EM structure of human islet amyloid polypeptide (hIAPP, or amylin) fibrils | | Descriptor: | Islet amyloid polypeptide (SUMO-tagged) | | Authors: | Cao, Q, Boyer, D.R, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2020-02-18 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure and inhibitor design of human IAPP (amylin) fibrils.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7M64
 
 | | Cryo-EM structure of human islet amyloid polypeptide (hIAPP, or amylin) fibrils seeded by patient extracted fibrils, polymorph 3 | | Descriptor: | Islet amyloid polypeptide | | Authors: | Cao, Q, Boyer, D.R, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2021-03-25 | | Release date: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of hIAPP fibrils seeded by patient-extracted fibrils reveal new polymorphs and conserved fibril cores
Nat.Struct.Mol.Biol., 28, 2021
|
|
7M61
 
 | | Cryo-EM structure of human islet amyloid polypeptide (hIAPP, or amylin) fibrils seeded by patient extracted fibrils, polymorph 1 | | Descriptor: | Islet amyloid polypeptide | | Authors: | Cao, Q, Boyer, D.R, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2021-03-25 | | Release date: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of hIAPP fibrils seeded by patient-extracted fibrils reveal new polymorphs and conserved fibril cores
Nat.Struct.Mol.Biol., 28, 2021
|
|
7M65
 
 | | Cryo-EM structure of human islet amyloid polypeptide (hIAPP, or amylin) fibrils seeded by patient extracted fibrils, polymorph 4 | | Descriptor: | Islet amyloid polypeptide | | Authors: | Cao, Q, Boyer, D.R, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2021-03-25 | | Release date: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of hIAPP fibrils seeded by patient-extracted fibrils reveal new polymorphs and conserved fibril cores
Nat.Struct.Mol.Biol., 28, 2021
|
|
7M62
 
 | | Cryo-EM structure of human islet amyloid polypeptide (hIAPP, or amylin) fibrils seeded by patient extracted fibrils, polymorph 2 | | Descriptor: | Islet amyloid polypeptide | | Authors: | Cao, Q, Boyer, D.R, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2021-03-25 | | Release date: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of hIAPP fibrils seeded by patient-extracted fibrils reveal new polymorphs and conserved fibril cores
Nat.Struct.Mol.Biol., 28, 2021
|
|
3V6M
 
 | | Inhibition of caspase-6 activity by single mutation outside the active site | | Descriptor: | Caspase-6 | | Authors: | Cao, Q, Wang, X.J, Liu, D.F, Li, L.F, Su, X.D. | | Deposit date: | 2011-12-20 | | Release date: | 2012-03-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.692 Å) | | Cite: | Inhibitory mechanism of caspase-6 phosphorylation revealed by crystal structures, molecular dynamics simulations, and biochemical assays
J.Biol.Chem., 287, 2012
|
|
3V6L
 
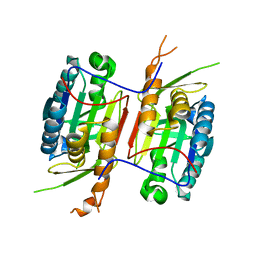 | | Crystal Structure of caspase-6 inactivation mutation | | Descriptor: | Caspase-6 | | Authors: | Cao, Q, Wang, X.J, Liu, D.F, Li, L.F, Su, X.D. | | Deposit date: | 2011-12-20 | | Release date: | 2012-03-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitory mechanism of caspase-6 phosphorylation revealed by crystal structures, molecular dynamics simulations, and biochemical assays
J.Biol.Chem., 287, 2012
|
|
4IYR
 
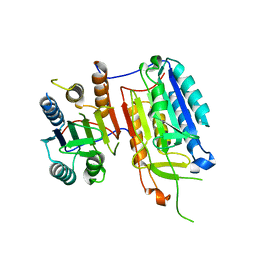 | | Crystal structure of full-length caspase-6 zymogen | | Descriptor: | Caspase-6 | | Authors: | Cao, Q, Wang, X.-J, Li, L.-F, Su, X.-D. | | Deposit date: | 2013-01-29 | | Release date: | 2014-01-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.697 Å) | | Cite: | The regulatory mechanism of the caspase 6 pro-domain revealed by crystal structure and biochemical assays.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6BCS
 
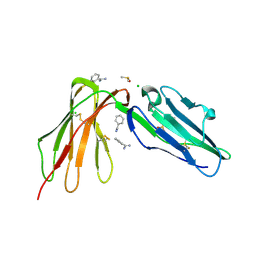 | | LilrB2 D1D2 domains complexed with benzamidine | | Descriptor: | BENZAMIDINE, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Cao, Q, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2017-10-20 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibiting amyloid-beta cytotoxicity through its interaction with the cell surface receptor LilrB2 by structure-based design.
Nat Chem, 10, 2018
|
|
6N3A
 
 | | SegA-long, conformation of TDP-43 low complexity domain segment A long | | Descriptor: | TAR DNA-binding protein 43, segA long small | | Authors: | Cao, Q, Boyer, D.R, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2018-11-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of four polymorphic TDP-43 amyloid cores.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6N37
 
 | | SegA-sym, conformation of TDP-43 low complexity domain segment A sym | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Cao, Q, Boyer, D.R, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2018-11-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of four polymorphic TDP-43 amyloid cores.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6N3C
 
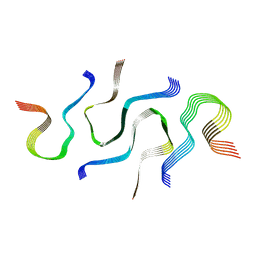 | | SegB, conformation of TDP-43 low complexity domain segment A | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Cao, Q, Boyer, D.R, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2018-11-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of four polymorphic TDP-43 amyloid cores.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6N3B
 
 | | SegA-asym, conformation of TDP-43 low complexity domain segment A asym | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Cao, Q, Boyer, D.R, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2018-11-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of four polymorphic TDP-43 amyloid cores.
Nat.Struct.Mol.Biol., 26, 2019
|
|
7SAQ
 
 | | Cryo-EM structure of TMEM106B fibrils extracted from a FTLD-TDP patient, polymorph 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | Authors: | Cao, Q, Jiang, Y, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2021-09-23 | | Release date: | 2022-03-09 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Amyloid fibrils in FTLD-TDP are composed of TMEM106B and not TDP-43.
Nature, 605, 2022
|
|
7SAR
 
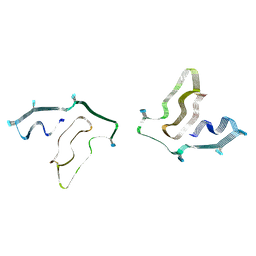 | | Cryo-EM structure of TMEM106B fibrils extracted from a FTLD-TDP patient, polymorph 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | Authors: | Cao, Q, Jiang, Y, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2021-09-23 | | Release date: | 2022-03-09 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Amyloid fibrils in FTLD-TDP are composed of TMEM106B and not TDP-43.
Nature, 605, 2022
|
|
7SAS
 
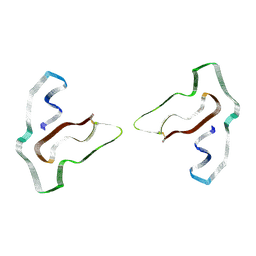 | | Cryo-EM structure of TMEM106B fibrils extracted from a FTLD-TDP patient, polymorph 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | Authors: | Cao, Q, Jiang, Y, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2021-09-23 | | Release date: | 2022-03-09 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Amyloid fibrils in FTLD-TDP are composed of TMEM106B and not TDP-43.
Nature, 605, 2022
|
|
8JMV
 
 | |
8JMW
 
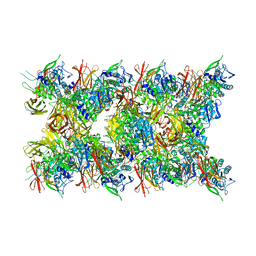 | | Fibril form of serine peptidase Vpr | | Descriptor: | S8 family serine peptidase | | Authors: | Cao, Q, Cheng, Y. | | Deposit date: | 2023-06-05 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Serine peptidase Vpr forms enzymatically active fibrils outside Bacillus bacteria revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
6WQK
 
 | | hnRNPA2 Low complexity domain (LCD) determined by cryoEM | | Descriptor: | MCherry fluorescent protein,Heterogeneous nuclear ribonucleoproteins A2/B1 chimera | | Authors: | Lu, J, Cao, Q, Hughes, M.P, Sawaya, M.R, Boyer, D.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2020-04-29 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | CryoEM structure of the low-complexity domain of hnRNPA2 and its conversion to pathogenic amyloid.
Nat Commun, 11, 2020
|
|
6WPQ
 
 | | GNYNVF from hnRNPA2-low complexity domain segment, residues 286-291, D290V variant | | Descriptor: | Heterogeneous nuclear ribonucleoprotein A2 | | Authors: | Lu, J, Cao, Q, Hughes, M.P, Sawaya, M.R, Boyer, D.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2020-04-27 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | CryoEM structure of the low-complexity domain of hnRNPA2 and its conversion to pathogenic amyloid.
Nat Commun, 11, 2020
|
|
3L87
 
 | |
3L9T
 
 | |
3OD5
 
 | | Crystal structure of active caspase-6 bound with Ac-VEID-CHO | | Descriptor: | CACODYLATE ION, Caspase-6, peptide aldehyde inhibitor AC-VEID-CHO | | Authors: | Wang, X.-J, Liu, X, Wang, K.-T, Cao, Q, Su, X.-D. | | Deposit date: | 2010-08-11 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of human caspase 6 reveal a new mechanism for intramolecular cleavage self-activation
Embo Rep., 11, 2010
|
|
6CB9
 
 | | Segment AALQSS from the low complexity domain of TDP-43, residues 328-333 | | Descriptor: | AALQSS | | Authors: | Guenther, E.L, Cao, Q, Lu, J, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2018-02-02 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6CEW
 
 | | Segment AMMAAA from the low complexity domain of TDP-43, residues 321-326 | | Descriptor: | AMMAAA | | Authors: | Guenther, E.L, Cao, Q, Lu, J, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2018-02-12 | | Release date: | 2018-04-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
