8SNX
 
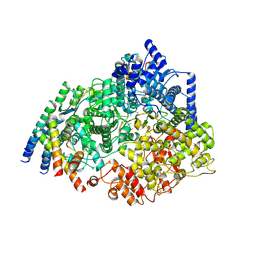 | | Cryo-EM structure of the respiratory syncytial virus polymerase (L:P) bound to the leader promoter | | Descriptor: | Phosphoprotein, RNA (5'-R(*UP*UP*UP*UP*UP*CP*GP*CP*GP*U)-3'), RNA-directed RNA polymerase L | | Authors: | Cao, D, Gao, Y, Chen, Z, Gooneratne, I, Roesler, C, Mera, C, Liang, B. | | Deposit date: | 2023-04-28 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of the promoter-bound respiratory syncytial virus polymerase.
Nature, 625, 2024
|
|
8SNY
 
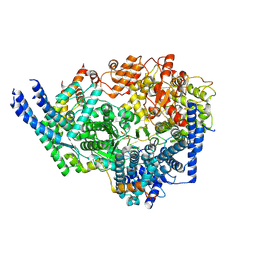 | | Cryo-EM structure of the respiratory syncytial virus polymerase (L:P) bound to the trailer complementary promoter | | Descriptor: | Phosphoprotein, RNA (5'-R(*UP*UP*UP*UP*UP*CP*UP*CP*GP*U)-3'), RNA-directed RNA polymerase L | | Authors: | Cao, D, Gao, Y, Chen, Z, Gooneratne, I, Roesler, C, Mera, C, Liang, B. | | Deposit date: | 2023-04-28 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structures of the promoter-bound respiratory syncytial virus polymerase.
Nature, 625, 2024
|
|
6UEN
 
 | |
8IHP
 
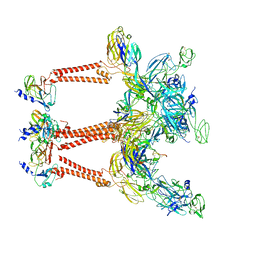 | | Structure of Semliki Forest virus VLP in complex with the receptor VLDLR-LA3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Capsid protein, ... | | Authors: | Cao, D, Ma, B, Cao, Z, Zhang, X, Xiang, Y. | | Deposit date: | 2023-02-23 | | Release date: | 2023-04-12 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of Semliki Forest virus in complex with its receptor VLDLR.
Cell, 186, 2023
|
|
5BTR
 
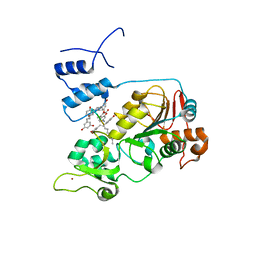 | | Crystal structure of SIRT1 in complex with resveratrol and an AMC-containing peptide | | Descriptor: | AMC-containing peptide, NAD-dependent protein deacetylase sirtuin-1, RESVERATROL, ... | | Authors: | Cao, D, Wang, M, Qiu, X, Liu, D, Jiang, H, Yang, N, Xu, R.M. | | Deposit date: | 2015-06-03 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for allosteric, substrate-dependent stimulation of SIRT1 activity by resveratrol
Genes Dev., 29, 2015
|
|
7CCR
 
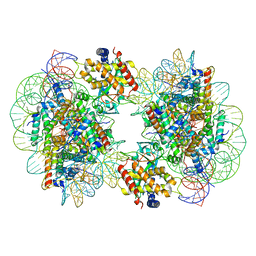 | | Structure of the 2:2 cGAS-nucleosome complex | | Descriptor: | Cyclic GMP-AMP synthase, DNA (147-MER), Histone H2A type 1-B/E, ... | | Authors: | Cao, D, Han, X, Fan, X, Xu, R.M, Zhang, X. | | Deposit date: | 2020-06-17 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis for nucleosome-mediated inhibition of cGAS activity.
Cell Res., 30, 2020
|
|
7CCQ
 
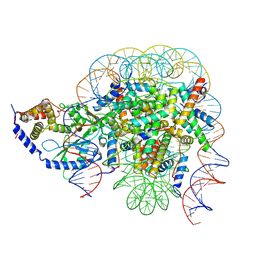 | | Structure of the 1:1 cGAS-nucleosome complex | | Descriptor: | Cyclic GMP-AMP synthase, DNA (147-MER), Histone H2A type 1-B/E, ... | | Authors: | Cao, D, Han, X, Fan, X, Xu, R.M, Zhang, X. | | Deposit date: | 2020-06-17 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for nucleosome-mediated inhibition of cGAS activity.
Cell Res., 30, 2020
|
|
7WC5
 
 | | Crystal structure of serotonin 2A receptor in complex with psilocin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[2-(dimethylamino)ethyl]-1~{H}-indol-4-ol, 5-hydroxytryptamine receptor 2A, ... | | Authors: | Cao, D, Yu, J, Wang, H, Luo, Z, Liu, X, He, L, Qi, J, Fan, L, Tang, L, Chen, Z, Li, J, Cheng, J, Wang, S. | | Deposit date: | 2021-12-18 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-based discovery of nonhallucinogenic psychedelic analogs.
Science, 375, 2022
|
|
7WC4
 
 | | Crystal structure of serotonin 2A receptor in complex with serotonin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2A,5-hydroxytryptamine receptor 2A,5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, CHOLESTEROL, ... | | Authors: | Cao, D, Yu, J, Wang, H, Luo, Z, Liu, X, He, L, Qi, J, Fan, L, Tang, L, Chen, Z, Li, J, Cheng, J, Wang, S. | | Deposit date: | 2021-12-18 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-based discovery of nonhallucinogenic psychedelic analogs.
Science, 375, 2022
|
|
7WC8
 
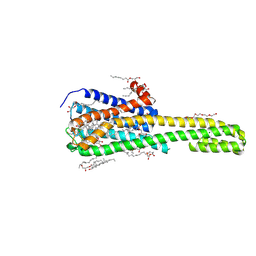 | | Crystal structure of serotonin 2A receptor in complex with lumateperone | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-(4-fluorophenyl)-4-[(10~{R},15~{S})-4-methyl-1,4,12-triazatetracyclo[7.6.1.0^{5,16}.0^{10,15}]hexadeca-5,7,9(16)-trien-12-yl]butan-1-one, 5-hydroxytryptamine receptor 2A,5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, ... | | Authors: | Cao, D, Yu, J, Wang, H, Luo, Z, Liu, X, He, L, Qi, J, Fan, L, Tang, L, Chen, Z, Li, J, Cheng, J, Wang, S. | | Deposit date: | 2021-12-18 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-based discovery of nonhallucinogenic psychedelic analogs.
Science, 375, 2022
|
|
7WC7
 
 | | Crystal structure of serotonin 2A receptor in complex with lisuride | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2A,5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, CHOLESTEROL, ... | | Authors: | Cao, D, Yu, J, Wang, H, Luo, Z, Liu, X, He, L, Qi, J, Fan, L, Tang, L, Chen, Z, Li, J, Cheng, J, Wang, S. | | Deposit date: | 2021-12-18 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based discovery of nonhallucinogenic psychedelic analogs.
Science, 375, 2022
|
|
7WC9
 
 | | Crystal structure of serotonin 2A receptor in complex with non-hallucinogenic psychedelic analog | | Descriptor: | (10~{R},15~{S})-12-[3-(2-methoxyphenyl)propyl]-4-methyl-1,4,12-triazatetracyclo[7.6.1.0^{5,16}.0^{10,15}]hexadeca-5(16),6,8-triene, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2A,5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, ... | | Authors: | Cao, D, Yu, J, Wang, H, Luo, Z, Liu, X, He, L, Qi, J, Fan, L, Tang, L, Chen, Z, Li, J, Cheng, J, Wang, S. | | Deposit date: | 2021-12-18 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based discovery of nonhallucinogenic psychedelic analogs.
Science, 375, 2022
|
|
7WC6
 
 | | Crystal structure of serotonin 2A receptor in complex with LSD | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (8alpha)-N,N-diethyl-6-methyl-9,10-didehydroergoline-8-carboxamide, 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, ... | | Authors: | Cao, D, Yu, J, Wang, H, Luo, Z, Liu, X, He, L, Qi, J, Fan, L, Tang, L, Chen, Z, Li, J, Cheng, J, Wang, S. | | Deposit date: | 2021-12-18 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based discovery of nonhallucinogenic psychedelic analogs.
Science, 375, 2022
|
|
8WIU
 
 | | Bromodomain and Extra-terminal Domain (BET) BRD4 | | Descriptor: | 7-[5-[1-(cyclopropylmethyl)-3,5-dimethyl-pyrazol-4-yl]pyridin-3-yl]-1~{H}-imidazo[4,5-b]pyridine, Isoform C of Bromodomain-containing protein 4 | | Authors: | Cao, D, Zhiyan, D, Xiong, B. | | Deposit date: | 2023-09-25 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of a brain-permeable bromodomain and extra terminal domain (BET) inhibitor with selectivity for BD1 for the treatment of multiple sclerosis.
Eur.J.Med.Chem., 265, 2023
|
|
8IBQ
 
 | | Bromodomain and Extra-terminal Domain (BET) BRD4 | | Descriptor: | 7-[2-fluoranyl-3-(1,3,5-trimethylpyrazol-4-yl)phenyl]-1~{H}-imidazo[4,5-b]pyridine, Bromodomain-containing protein 4 | | Authors: | Cao, D, Zhiyan, D, Xiong, B. | | Deposit date: | 2023-02-10 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of 1 H -Imidazo[4,5- b ]pyridine Derivatives as Potent and Selective BET Inhibitors for the Management of Neuropathic Pain.
J.Med.Chem., 66, 2023
|
|
8IDH
 
 | | Bromodomain and Extra-terminal Domain (BET) BRD4 | | Descriptor: | 7-[2-fluoranyl-3-(1,3,5-trimethylpyrazol-4-yl)phenyl]-1~{H}-imidazo[4,5-b]pyridine, Bromodomain-containing protein 4 | | Authors: | Cao, D, Zhiyan, D, Xiong, B. | | Deposit date: | 2023-02-13 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Discovery of 1 H -Imidazo[4,5- b ]pyridine Derivatives as Potent and Selective BET Inhibitors for the Management of Neuropathic Pain.
J.Med.Chem., 66, 2023
|
|
7X6T
 
 | | Discovery of Selective BRD4 BD1 Inhibitor Based on [1,2,4] triazolo [4,3-b] pyridazine Scaffold | | Descriptor: | (2~{R})-2-[[3-methyl-6-(2-phenoxyphenyl)-[1,2,4]triazolo[4,3-b]pyridazin-8-yl]amino]propanamide, Isoform C of Bromodomain-containing protein 4 | | Authors: | Cao, D, Xiong, B. | | Deposit date: | 2022-03-08 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Discovery of Selective BRD4 BD1 Inhibitor Based on [1,2,4] triazolo [4,3-b] pyridazine Scaffold
To Be Published
|
|
7XY3
 
 | |
7XY4
 
 | |
6PZQ
 
 | |
7DNO
 
 | | Characterization of Peptide Ligands Against WDR5 Isolated Using Phage Display Technique | | Descriptor: | CYS-ARG-THR-LEU-PRO-PHE, WD repeat-containing protein 5 | | Authors: | Cao, J, Cao, D, Xiong, B, Li, Y, Fan, T. | | Deposit date: | 2020-12-10 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Phage-Display Based Discovery and Characterization of Peptide Ligands against WDR5.
Molecules, 26, 2021
|
|
5X58
 
 | | Prefusion structure of SARS-CoV spike glycoprotein, conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X5B
 
 | | Prefusion structure of SARS-CoV spike glycoprotein, conformation 2 | | Descriptor: | Spike glycoprotein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2017-05-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5XI4
 
 | | BRD4 bound with compound Bdi4 | | Descriptor: | (3~{S})-4-cyclopropyl-1,3-dimethyl-6-[[(1~{S})-1-(4-methylphenyl)ethyl]amino]-3~{H}-quinoxalin-2-one, Bromodomain-containing protein 4 | | Authors: | Xiong, B, Cao, D, Li, Y. | | Deposit date: | 2017-04-25 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.486 Å) | | Cite: | BRD4 bound with compound Bdi4
To Be Published
|
|
5X59
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, three-fold symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
