2A79
 
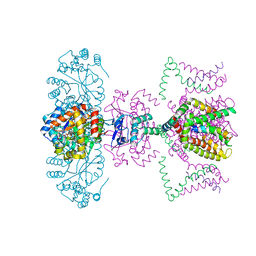 | | Mammalian Shaker Kv1.2 potassium channel- beta subunit complex | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, Potassium voltage-gated channel subfamily A member 2, ... | | Authors: | Long, S.B, Campbell, E.B, MacKinnon, R. | | Deposit date: | 2005-07-05 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a mammalian voltage-dependent Shaker family K+ channel.
Science, 309, 2005
|
|
2R9R
 
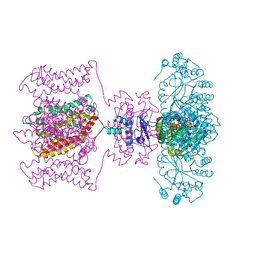 | | Shaker family voltage dependent potassium channel (kv1.2-kv2.1 paddle chimera channel) in association with beta subunit | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, ... | | Authors: | Long, S.B, Tao, X, Campbell, E.B, Mackinnon, R. | | Deposit date: | 2007-09-13 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment.
Nature, 450, 2007
|
|
1OTS
 
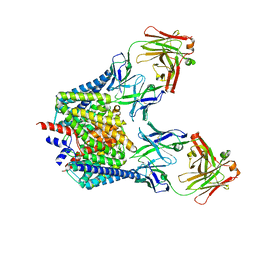 | | Structure of the Escherichia coli ClC Chloride channel and Fab Complex | | Descriptor: | CHLORIDE ION, Fab fragment (heavy chain), Fab fragment (light chain), ... | | Authors: | Dutzler, R, Campbell, E.B, MacKinnon, R. | | Deposit date: | 2003-03-22 | | Release date: | 2003-04-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Gating the Selectivity Filter in ClC Chloride Channels
Science, 300, 2003
|
|
1OTU
 
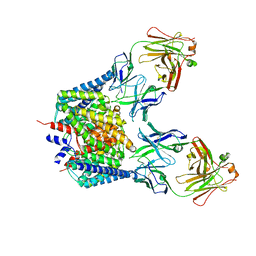 | | Structure of the Escherichia coli ClC Chloride channel E148Q mutant and Fab Complex | | Descriptor: | CHLORIDE ION, Fab fragment (Heavy chain), Fab fragment (Light chain), ... | | Authors: | Dutzler, R, Campbell, E.B, MacKinnon, R. | | Deposit date: | 2003-03-23 | | Release date: | 2003-04-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Gating the Selectivity Filter in ClC Chloride Channels
Science, 300, 2003
|
|
1OTT
 
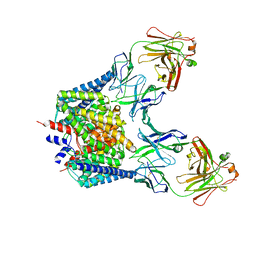 | | Structure of the Escherichia coli ClC Chloride channel E148A mutant and Fab Complex | | Descriptor: | CHLORIDE ION, Fab fragment (Heavy chain), Fab fragment (Light chain), ... | | Authors: | Dutzler, R, Campbell, E.B, MacKinnon, R. | | Deposit date: | 2003-03-23 | | Release date: | 2003-04-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Gating the Selectivity Filter in ClC Chloride Channels
Science, 300, 2003
|
|
1KPK
 
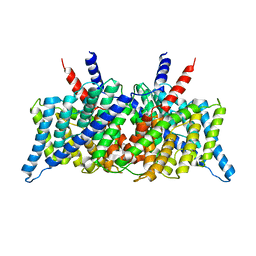 | | Crystal Structure of the ClC Chloride Channel from E. coli | | Descriptor: | putative channel transporter | | Authors: | Dutzler, R, Campbell, E.B, Cadene, M, Chait, B.T, MacKinnon, R. | | Deposit date: | 2001-12-31 | | Release date: | 2002-01-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | X-ray structure of a ClC chloride channel at 3.0 A reveals the molecular basis of
anion selectivity.
Nature, 415, 2002
|
|
1KPL
 
 | | Crystal Structure of the ClC Chloride Channel from S. typhimurium | | Descriptor: | CHLORIDE ION, N-OCTANE, PENTADECANE, ... | | Authors: | Dutzler, R, Campbell, E.B, Cadene, M, Chait, B.T, MacKinnon, R. | | Deposit date: | 2001-12-31 | | Release date: | 2002-01-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structure of a ClC chloride channel at 3.0 A reveals the molecular basis of
anion selectivity.
Nature, 415, 2002
|
|
3LUT
 
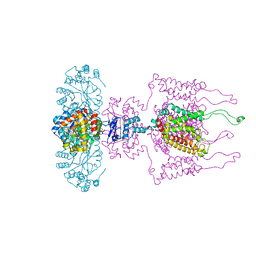 | | A Structural Model for the Full-length Shaker Potassium Channel Kv1.2 | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, Potassium voltage-gated channel subfamily A member 2, ... | | Authors: | Chen, X, Ni, F, Wang, Q, Ma, J. | | Deposit date: | 2010-02-18 | | Release date: | 2010-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the full-length Shaker potassium channel Kv1.2 by normal-mode-based X-ray crystallographic refinement.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4WFH
 
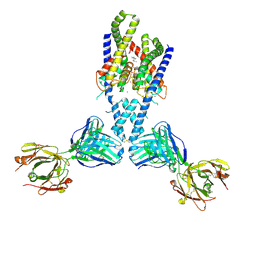 | |
4WFG
 
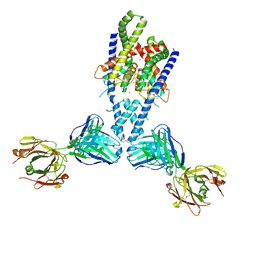 | | Human TRAAK K+ channel in a Tl+ bound conductive conformation | | Descriptor: | ANTI-TRAAK ANTIBODY 13E9 FAB FRAGMENT HEAVY CHAIN, ANTI-TRAAK ANTIBODY 13E9 FAB FRAGMENT LIGHT CHAIN, CALCIUM ION, ... | | Authors: | Brohawn, S.G, MacKinnon, R. | | Deposit date: | 2014-09-15 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Physical mechanism for gating and mechanosensitivity of the human TRAAK K+ channel.
Nature, 516, 2014
|
|
4WFE
 
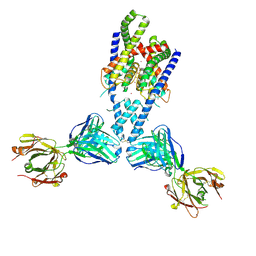 | | Human TRAAK K+ channel in a K+ bound conductive conformation | | Descriptor: | ANTI-TRAAK ANTIBODY 13E9 FAB FRAGMENT HEAVY CHAIN, ANTI-TRAAK ANTIBODY 13E9 FAB FRAGMENT LIGHT CHAIN, CALCIUM ION, ... | | Authors: | Brohawn, S.G, MacKinnon, R. | | Deposit date: | 2014-09-15 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Physical mechanism for gating and mechanosensitivity of the human TRAAK K+ channel.
Nature, 516, 2014
|
|
4WFF
 
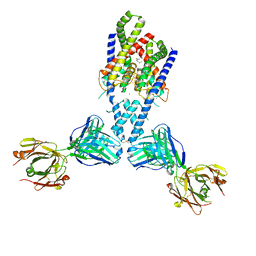 | |
4FG6
 
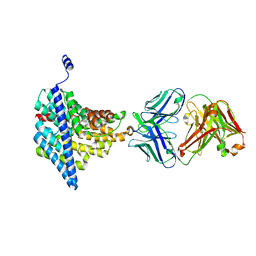 | | Structure of EcCLC E148A mutant in Glutamate | | Descriptor: | Fab fragment (Heavy chain), Fab fragment (Light chain), H(+)/Cl(-) exchange transporter ClcA | | Authors: | Feng, L, MacKinnon, R. | | Deposit date: | 2012-06-04 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.019 Å) | | Cite: | Molecular mechanism of proton transport in CLC Cl-/H+ exchange transporters.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6PIS
 
 | |
5TQQ
 
 | |
5TR1
 
 | |
4I9W
 
 | |
1BLD
 
 | | BASIC FIBROBLAST GROWTH FACTOR (FGF-2) MUTANT WITH CYS 78 REPLACED BY SER AND CYS 96 REPLACED BY SER, NMR | | Descriptor: | BASIC FIBROBLAST GROWTH FACTOR | | Authors: | Powers, R, Seddon, A.P, Bohlen, P, Moy, F.J. | | Deposit date: | 1996-05-20 | | Release date: | 1996-11-08 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of basic fibroblast growth factor determined by multidimensional heteronuclear magnetic resonance spectroscopy.
Biochemistry, 35, 1996
|
|
3ORG
 
 | |
1BLA
 
 | | BASIC FIBROBLAST GROWTH FACTOR (FGF-2) MUTANT WITH CYS 78 REPLACED BY SER AND CYS 96 REPLACED BY SER, NMR | | Descriptor: | BASIC FIBROBLAST GROWTH FACTOR | | Authors: | Powers, R, Seddon, A.P, Bohlen, P, Moy, F.J. | | Deposit date: | 1996-05-20 | | Release date: | 1996-11-08 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of basic fibroblast growth factor determined by multidimensional heteronuclear magnetic resonance spectroscopy.
Biochemistry, 35, 1996
|
|
