5W7H
 
 | | Supercharged arPTE variant R5 | | Descriptor: | Phosphotriesterase, ZINC ION | | Authors: | Campbell, E, Grant, J, Wang, Y, Sandhu, M, Williams, R.J, Nisbet, D.R, Perriman, A, Lupton, D, Jackson, C.J. | | Deposit date: | 2017-06-19 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Hydrogel-Immobilized Supercharged Proteins
Adv Biosyst, 2018
|
|
4PBE
 
 | | Phosphotriesterase Variant Rev6 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-revR6, ... | | Authors: | Campbell, E, Kaltenbach, M, Tokuriki, N, Jackson, C.J. | | Deposit date: | 2014-04-12 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat. Chem. Biol., 12, 2016
|
|
4PBF
 
 | | Phosphotriesterase variant Rev12 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-revR12, ... | | Authors: | Campbell, E, Kaltenbach, M, Tokuriki, N, Jackson, C.J. | | Deposit date: | 2014-04-12 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat. Chem. Biol., 12, 2016
|
|
4PCP
 
 | | Crystal structure of Phosphotriesterase variant R0 | | Descriptor: | CACODYLATE ION, Phosphotriesterase variant PTE-R0, ZINC ION | | Authors: | Campbell, E, Kaltenbach, M, Tokuriki, N, Jackson, C.J. | | Deposit date: | 2014-04-16 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat. Chem. Biol., 12, 2016
|
|
4XD5
 
 | | Phosphotriesterase variant R2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-R2, ... | | Authors: | Campbell, E, Kaltenbach, M, Tokuriki, N, Jackson, C.J. | | Deposit date: | 2014-12-19 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
4XAY
 
 | | Cycles of destabilization and repair underlie evolutionary transitions in enzymes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-R8, ... | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-16 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
4XAZ
 
 | | Cycles of destabilization and repair underlie evolutionary transitions in enzymes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Phosphotriesterase variant PTE-R18, ZINC ION | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-16 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
4XD4
 
 | | Phosphotriesterase variant E2b | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-R3, ... | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-19 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
4XAG
 
 | | Cycles of destabilization and repair underlie the evolution of new enzyme function | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-R6, ... | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-14 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
4XD3
 
 | | Phosphotriesterase variant E3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-E1, ... | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-19 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
4PCN
 
 | | Phosphotriesterase variant R22 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Phosphotriesterase variant PTE-R22, ZINC ION | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-04-16 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat. Chem. Biol., 12, 2016
|
|
4XD6
 
 | | Phosphotriesterase Variant E2a | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-E2, ... | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-19 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
4XAF
 
 | | Cycles of destabilization and repair underlie evolutionary transitions in enzymes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-R1, ... | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-14 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
7QE2
 
 | | Crystal structure of D-glucuronic acid bound to SN243 | | Descriptor: | ACETATE ION, SN243, SULFATE ION, ... | | Authors: | Neun, S, Brear, P, Campbell, E, Omari, K, Wagner, O, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2021-12-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Functional metagenomic screening identifies an unexpected beta-glucuronidase.
Nat.Chem.Biol., 18, 2022
|
|
7QG4
 
 | | Apo crystal structure of a mutant of SN243 (D415N) | | Descriptor: | SN243, SULFATE ION, ZINC ION | | Authors: | Neun, S, Brear, P, Campbell, E, Omari, K, Wagner, O, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2021-12-07 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Functional metagenomic screening identifies an unexpected beta-glucuronidase.
Nat.Chem.Biol., 18, 2022
|
|
7QEF
 
 | | Crystal structure of para-nitrophenyl-Beta-D-glucuronide bound to a mutant of SN243 (D415A) | | Descriptor: | 4-nitrophenyl beta-D-glucopyranosiduronic acid, ACETATE ION, SN243, ... | | Authors: | Neun, S, Brear, P, Campbell, E, Omari, K, Wagner, O, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2021-12-02 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Functional metagenomic screening identifies an unexpected beta-glucuronidase.
Nat.Chem.Biol., 18, 2022
|
|
7QEA
 
 | | Crystal structure of fluorescein-di-Beta-D-glucuronide bound to a mutant of SN243 (D415A) | | Descriptor: | (2~{S},3~{S},4~{S},5~{R},6~{S})-3,4,5-tris(oxidanyl)-6-[(1~{R})-6'-oxidanyl-3-oxidanylidene-spiro[2-benzofuran-1,9'-xanthene]-3'-yl]oxy-oxane-2-carboxylic acid, ACETATE ION, SN243, ... | | Authors: | Neun, S, Brear, P, Campbell, E, Omari, K, Wagner, O, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2021-12-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Functional metagenomic screening identifies an unexpected beta-glucuronidase.
Nat.Chem.Biol., 18, 2022
|
|
7QE1
 
 | | Crystal structure of apo SN243 | | Descriptor: | SN243, ZINC ION | | Authors: | Neun, S, Brear, P, Campbell, E, Omari, K, Wagner, O, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2021-12-01 | | Release date: | 2022-10-12 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Functional metagenomic screening identifies an unexpected beta-glucuronidase.
Nat.Chem.Biol., 18, 2022
|
|
7QEE
 
 | | SN243 mutant D415N bound to para-nitrophenyl-Beta-D-glucuronide | | Descriptor: | 4-nitrophenyl beta-D-glucopyranosiduronic acid, SN243, SULFATE ION, ... | | Authors: | Neun, S, Brear, P, Campbell, E, Omari, K, Wagner, O, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2021-12-02 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.374 Å) | | Cite: | Functional metagenomic screening identifies an unexpected beta-glucuronidase.
Nat.Chem.Biol., 18, 2022
|
|
2FMN
 
 | | Ala177Val mutant of E. coli Methylenetetrahydrofolate Reductase complex with LY309887 | | Descriptor: | 5,10-methylenetetrahydrofolate reductase, FLAVIN-ADENINE DINUCLEOTIDE, N-[(5-{2-[(6R)-2-AMINO-4-OXO-3,4,5,6,7,8-HEXAHYDROPYRIDO[2,3-D]PYRIMIDIN-6-YL]ETHYL}-2-THIENYL)CARBONYL]-L-GLUTAMIC ACID | | Authors: | Pejchal, R, Campbell, E, Guenther, B.D, Lennon, B.W, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 2006-01-09 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Perturbations in the Ala -> Val Polymorphism of Methylenetetrahydrofolate Reductase: How Binding of Folates May Protect against Inactivation
Biochemistry, 45, 2006
|
|
2FMO
 
 | | Ala177Val mutant of E. coli Methylenetetrahydrofolate Reductase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5,10-methylenetetrahydrofolate reductase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Pejchal, R, Campbell, E, Guenther, B.D, Lennon, B.W, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 2006-01-09 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Perturbations in the Ala -> Val Polymorphism of Methylenetetrahydrofolate Reductase: How Binding of Folates May Protect against Inactivation
Biochemistry, 45, 2006
|
|
4JTA
 
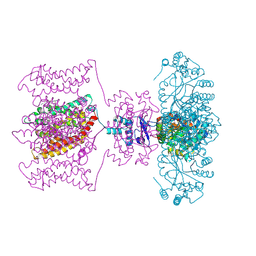 | | Crystal structure of Kv1.2-2.1 paddle chimera channel in complex with Charybdotoxin | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, ... | | Authors: | MacKinnon, R, Banerjee, A, Lee, A, Campbell, E. | | Deposit date: | 2013-03-23 | | Release date: | 2013-06-12 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a pore-blocking toxin in complex with a eukaryotic voltage-dependent K(+) channel.
Elife, 2, 2013
|
|
4JTD
 
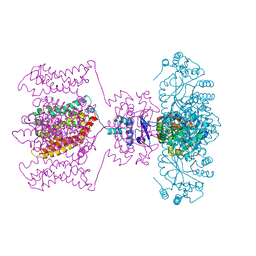 | | Crystal structure of Kv1.2-2.1 paddle chimera channel in complex with Lys27Met mutant of Charybdotoxin | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, ... | | Authors: | Banerjee, A, Lee, A, Campbell, E, MacKinnon, R. | | Deposit date: | 2013-03-23 | | Release date: | 2013-06-12 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structure of a pore-blocking toxin in complex with a eukaryotic voltage-dependent K(+) channel.
Elife, 2, 2013
|
|
4JTC
 
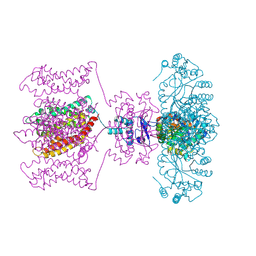 | | Crystal structure of Kv1.2-2.1 paddle chimera channel in complex with Charybdotoxin in Cs+ | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Banerjee, A, Lee, A, Campbell, E, MacKinnon, R. | | Deposit date: | 2013-03-23 | | Release date: | 2013-06-12 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structure of a pore-blocking toxin in complex with a eukaryotic voltage-dependent K(+) channel.
Elife, 2, 2013
|
|
5AF0
 
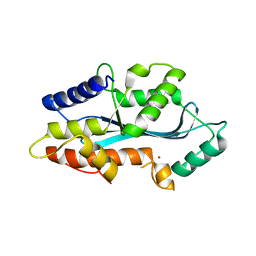 | | MAEL domain from Bombyx mori Maelstrom | | Descriptor: | MAELSTROM, ZINC ION | | Authors: | Chen, K, Campbell, E, Pandey, R.R, Yang, Z, McCarthy, A.A, Pillai, R.S. | | Deposit date: | 2015-01-13 | | Release date: | 2015-04-01 | | Last modified: | 2017-03-15 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Metazoan Maelstrom is an RNA-Binding Protein that Has Evolved from an Ancient Nuclease Active in Protists.
RNA, 21, 2015
|
|
