5OVO
 
 | |
5OYJ
 
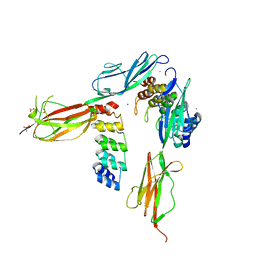 | | Crystal structure of VEGFR-2 domains 4-5 in complex with DARPin D4b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CACODYLATE ION, ... | | Authors: | Piscitelli, C.L, Thieltges, K.M, Markovic-Mueller, S, Binz, H.K, Ballmer-Hofer, K. | | Deposit date: | 2017-09-10 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Characterization of a drug-targetable allosteric site regulating vascular endothelial growth factor signaling.
Angiogenesis, 21, 2018
|
|
5T3W
 
 | |
5T3T
 
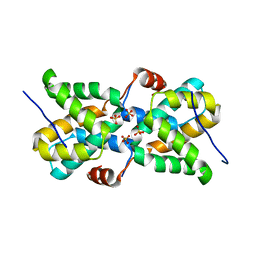 | | Ebola virus VP30 CTD bound to nucleoprotein | | Descriptor: | Fusion protein of Nucleoprotein and Minor nucleoprotein VP30, SULFATE ION | | Authors: | Kirchdoerfer, R.N, Moyer, C.L, Abelson, D.M, Saphire, E.O. | | Deposit date: | 2016-08-26 | | Release date: | 2016-09-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Ebola Virus VP30-NP Interaction Is a Regulator of Viral RNA Synthesis.
Plos Pathog., 12, 2016
|
|
5T6P
 
 | |
5T5X
 
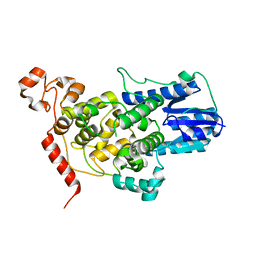 | |
5T78
 
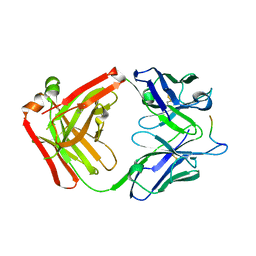 | | Crystal structure of therapeutic mAB AR20.5 in complex with MUC1 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, Fab Fragment - AR20.5 - Heavy chain, Fab fragment AR20.5 - Light Chain, ... | | Authors: | Brooks, C.L, Movahedin, M. | | Deposit date: | 2016-09-02 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Glycosylation of MUC1 influences the binding of a therapeutic antibody by altering the conformational equilibrium of the antigen.
Glycobiology, 27, 2017
|
|
5T6O
 
 | |
1CZN
 
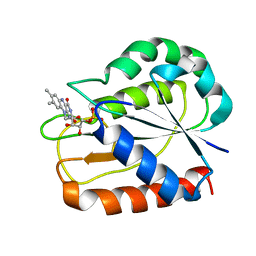 | | REFINED STRUCTURES OF OXIDIZED FLAVODOXIN FROM ANACYSTIS NIDULANS | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Smith, W.W, Pattridge, K.A, Luschinsky, C.L, Ludwig, M.L. | | Deposit date: | 1999-09-03 | | Release date: | 1999-12-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Refined structures of oxidized flavodoxin from Anacystis nidulans.
J.Mol.Biol., 294, 1999
|
|
1CZL
 
 | | COMPARISONS OF WILD TYPE AND MUTANT FLAVODOXINS FROM ANACYSTIS NIDULANS. STRUCTURAL DETERMINANTS OF THE REDOX POTENTIALS. | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Hoover, D.M, Drennan, C.L, Metzger, A.L, Osborne, C, Weber, C.H, Pattridge, K.A, Ludwig, M.L. | | Deposit date: | 1999-09-03 | | Release date: | 1999-12-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparisons of wild-type and mutant flavodoxins from Anacystis nidulans. Structural determinants of the redox potentials.
J.Mol.Biol., 294, 1999
|
|
1CZH
 
 | | COMPARISONS OF WILD TYPE AND MUTANT FLAVODOXINS FROM ANACYSTIS NIDULANS. STRUCTURAL DETERMINANTS OF THE REDOX POTENTIALS. | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN, GLYCEROL, ... | | Authors: | Hoover, D.M, Drennan, C.L, Metzger, A.L, Osborne, C, Weber, C.H, Pattridge, K.A, Ludwig, M.L. | | Deposit date: | 1999-09-03 | | Release date: | 1999-12-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Comparisons of wild-type and mutant flavodoxins from Anacystis nidulans. Structural determinants of the redox potentials.
J.Mol.Biol., 294, 1999
|
|
1CZR
 
 | | COMPARISONS OF WILD TYPE AND MUTANT FLAVODOXINS FROM ANACYSTIS NIDULANS. STRUCTURAL DETERMINANTS OF THE REDOX POTENTIALS. | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Hoover, D.M, Drennan, C.L, Metzger, A.L, Osborne, C, Weber, C.H, Pattridge, K.A, Ludwig, M.L. | | Deposit date: | 1999-09-07 | | Release date: | 1999-12-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparisons of wild-type and mutant flavodoxins from Anacystis nidulans. Structural determinants of the redox potentials.
J.Mol.Biol., 294, 1999
|
|
1D04
 
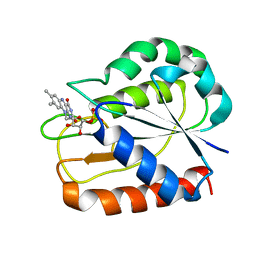 | | COMPARISONS OF WILD TYPE AND MUTANT FLAVODOXINS FROM ANACYSTIS NIDULANS. STRUCTURAL DETERMINANTS OF THE REDOX POTENTIALS. | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Hoover, D.M, Drennan, C.L, Metzger, A.L, Osborne, C, Weber, C.H, Pattridge, K.A, Ludwig, M.L. | | Deposit date: | 1999-09-08 | | Release date: | 1999-12-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Comparisons of wild-type and mutant flavodoxins from Anacystis nidulans. Structural determinants of the redox potentials.
J.Mol.Biol., 294, 1999
|
|
1CZO
 
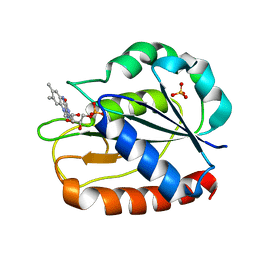 | | COMPARISONS OF WILD TYPE AND MUTANT FLAVODOXINS FROM ANACYSTIS NIDULANS. STRUCTURAL DETERMINANTS OF THE REDOX POTENTIALS. | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN, SULFATE ION | | Authors: | Hoover, D.M, Drennan, C.L, Metzger, A.L, Osborne, C, Weber, C.H, Pattridge, K.A, Ludwig, M.L. | | Deposit date: | 1999-09-03 | | Release date: | 1999-12-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Comparisons of wild-type and mutant flavodoxins from Anacystis nidulans. Structural determinants of the redox potentials.
J.Mol.Biol., 294, 1999
|
|
1CZK
 
 | | COMPARISONS OF WILD TYPE AND MUTANT FLAVODOXINS FROM ANACYSTIS NIDULANS. STRUCTURAL DETERMINANTS OF THE REDOX POTENTIALS. | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Hoover, D.M, Drennan, C.L, Metzger, A.L, Osborne, C, Weber, C.H, Pattridge, K.A, Ludwig, M.L. | | Deposit date: | 1999-09-03 | | Release date: | 2000-12-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparisons of wild-type and mutant flavodoxins from Anacystis nidulans. Structural determinants of the redox potentials.
J.Mol.Biol., 294, 1999
|
|
4V9I
 
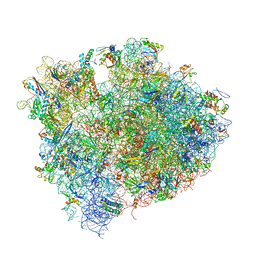 | | Crystal structure of thermus thermophilus 70S in complex with tRNAs and mRNA containing a pseudouridine in a stop codon | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S Ribosomal protein S10, ... | | Authors: | Fernandez, I.S, Ng, C.L, Kelley, A.C, Guowei, W, Yu, Y.T, Ramakrishnan, V. | | Deposit date: | 2013-04-04 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Unusual base pairing during the decoding of a stop codon by the ribosome.
Nature, 500, 2013
|
|
4V4M
 
 | | 1.45 Angstrom Structure of STNV coat protein | | Descriptor: | CALCIUM ION, Coat protein | | Authors: | Lane, S.W, Dennis, C.A, Lane, C.L, Trinh, C.H, Rizkallah, P.J, Stockley, P.G, Phillips, S.E.V. | | Deposit date: | 2011-04-28 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Construction and crystal structure of recombinant STNV capsids.
J.Mol.Biol., 413, 2011
|
|
4V5K
 
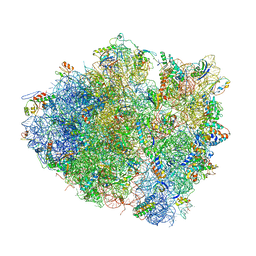 | | Structure of cytotoxic domain of colicin E3 bound to the 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ng, C.L, Lang, K, Meenan, N.A.G, Sharma, A, Kelley, A.C, Kleanthous, C, Ramakrishnan, V. | | Deposit date: | 2010-05-29 | | Release date: | 2014-07-09 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis for 16S Ribosomal RNA Cleavage by the Cytotoxic Domain of Colicin E3.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4XC7
 
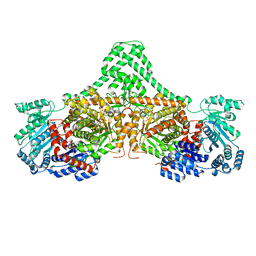 | |
4XYN
 
 | | X-ray structure of Ca(2+)-S100B with human RAGE-derived W61 peptide | | Descriptor: | CALCIUM ION, Protein S100-B, Receptor for advanced glycation endproducts-derived peptide (W61) | | Authors: | Jensen, J.L, Indurthi, V.S.K, Neau, D, Vetter, S.W, Colbert, C.L. | | Deposit date: | 2015-02-02 | | Release date: | 2015-05-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural insights into the binding of the human receptor for advanced glycation end products (RAGE) by S100B, as revealed by an S100B-RAGE-derived peptide complex.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XC6
 
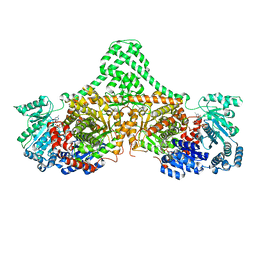 | | Isobutyryl-CoA mutase fused with bound adenosylcobalamin, GDP, and Mg (holo-IcmF/GDP) | | Descriptor: | 5'-DEOXYADENOSINE, ACETATE ION, COBALAMIN, ... | | Authors: | Jost, M, Drennan, C.L. | | Deposit date: | 2014-12-17 | | Release date: | 2015-02-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Visualization of a radical B12 enzyme with its G-protein chaperone.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8D02
 
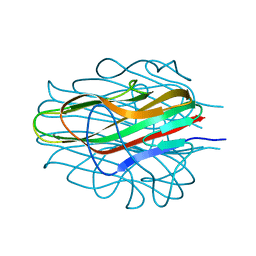 | | Crystal Structure of Bacillus anthracis BxpB | | Descriptor: | BxpB, CALCIUM ION | | Authors: | Chattopadhyay, D, Turnbough, C.L. | | Deposit date: | 2022-05-25 | | Release date: | 2023-05-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure and induced stability of trimeric BxpB: implications for the assembly of BxpB-BclA complexes in the exosporium of Bacillus anthracis.
Mbio, 14, 2023
|
|
1BMT
 
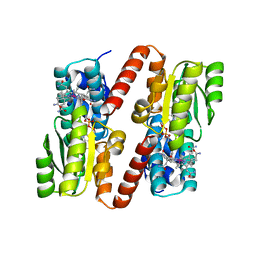 | | HOW A PROTEIN BINDS B12: A 3.O ANGSTROM X-RAY STRUCTURE OF THE B12-BINDING DOMAINS OF METHIONINE SYNTHASE | | Descriptor: | CO-METHYLCOBALAMIN, METHIONINE SYNTHASE | | Authors: | Drennan, C.L, Huang, S, Drummond, J.T, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 1994-09-02 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | How a protein binds B12: A 3.0 A X-ray structure of B12-binding domains of methionine synthase.
Science, 266, 1994
|
|
1CVX
 
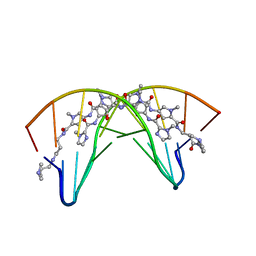 | | CRYSTAL STRUCTURE OF POLYAMIDE DIMER (IMPYHPPYBETADP)2 BOUND TO B-DNA DECAMER CCAGATCTGG | | Descriptor: | 5'-D(*CP*CP*AP*GP*AP*TP*CP*TP*GP*G)-3', HYDROXYPYRROLE-IMIDAZOLE-PYRROLE POLYAMIDE | | Authors: | Kielkopf, C.L, Bremer, R.E, White, S, Baird, E.E, Dervan, P.B, Rees, D.C. | | Deposit date: | 1999-08-24 | | Release date: | 2000-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural effects of DNA sequence on T.A recognition by hydroxypyrrole/pyrrole pairs in the minor groove.
J.Mol.Biol., 295, 2000
|
|
1CVY
 
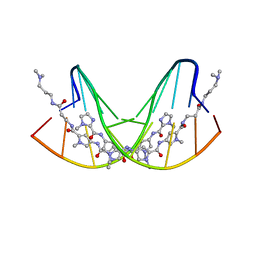 | | CRYSTAL STRUCTURE OF POLYAMIDE DIMER (IMPYPYPYBETADP)2 BOUND TO CCAGATCTGG | | Descriptor: | 5'-D(*CP*CP*AP*GP*AP*TP*CP*TP*GP*G)-3', IMIDAZOLE-PYRROLE POLYAMIDE | | Authors: | Kielkopf, C.L, Bremer, R.E, White, S, Baird, E.E, Dervan, P.B, Rees, D.C. | | Deposit date: | 1999-08-24 | | Release date: | 2000-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural effects of DNA sequence on T.A recognition by hydroxypyrrole/pyrrole pairs in the minor groove.
J.Mol.Biol., 295, 2000
|
|
