3H8K
 
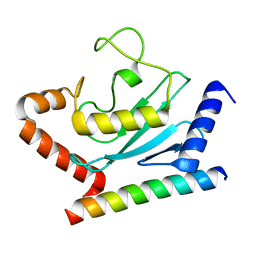 | | Crystal structure of Ube2g2 complxed with the G2BR domain of gp78 at 1.8-A resolution | | Descriptor: | Autocrine motility factor receptor, isoform 2, Ubiquitin-conjugating enzyme E2 G2 | | Authors: | Kalathur, R.C, Das, R, Li, J, Byrd, R.A, Ji, X. | | Deposit date: | 2009-04-29 | | Release date: | 2009-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Allosteric activation of E2-RING finger-mediated ubiquitylation by a structurally defined specific E2-binding region of gp78.
Mol.Cell, 34, 2009
|
|
1GA3
 
 | |
1NK1
 
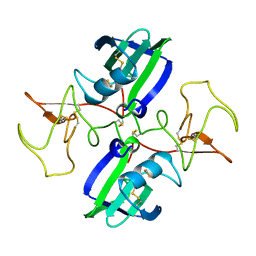 | | NK1 FRAGMENT OF HUMAN HEPATOCYTE GROWTH FACTOR/SCATTER FACTOR (HGF/SF) AT 2.5 ANGSTROM RESOLUTION | | Descriptor: | PROTEIN (HEPATOCYTE GROWTH FACTOR PRECURSOR) | | Authors: | Chirgadze, D.Y, Hepple, J.P, Zhou, H, Byrd, R.A, Blundell, T.L, Gherardi, E. | | Deposit date: | 1998-08-20 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the NK1 fragment of HGF/SF suggests a novel mode for growth factor dimerization and receptor binding.
Nat.Struct.Biol., 6, 1999
|
|
5HCK
 
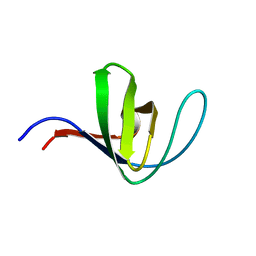 | | HUMAN HCK SH3 DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | HEMATOPOIETIC CELL KINASE | | Authors: | Horita, D.A, Baldisseri, D.M, Zhang, W, Altieri, A.S, Smithgall, T.E, Gmeiner, W.H, Byrd, R.A. | | Deposit date: | 1998-03-09 | | Release date: | 1998-06-17 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the human Hck SH3 domain and identification of its ligand binding site.
J.Mol.Biol., 278, 1998
|
|
4LAD
 
 | | Crystal Structure of the Ube2g2:RING-G2BR complex | | Descriptor: | E3 ubiquitin-protein ligase AMFR, OXALATE ION, Ubiquitin-conjugating enzyme E2 G2, ... | | Authors: | Liang, Y.-H, Li, J, Das, R, Byrd, R.A, Ji, X. | | Deposit date: | 2013-06-19 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine.
Embo J., 32, 2013
|
|
4HCK
 
 | | HUMAN HCK SH3 DOMAIN, NMR, 25 STRUCTURES | | Descriptor: | HEMATOPOIETIC CELL KINASE | | Authors: | Horita, D.A, Baldisseri, D.M, Zhang, W, Altieri, A.S, Smithgall, T.E, Gmeiner, W.H, Byrd, R.A. | | Deposit date: | 1998-03-09 | | Release date: | 1998-06-17 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the human Hck SH3 domain and identification of its ligand binding site.
J.Mol.Biol., 278, 1998
|
|
2HGF
 
 | | HAIRPIN LOOP CONTAINING DOMAIN OF HEPATOCYTE GROWTH FACTOR, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | HEPATOCYTE GROWTH FACTOR | | Authors: | Zhou, H, Mazzulla, M.J, Kaufman, J.D, Stahl, S.J, Wingfield, P.T, Rubin, J.S, Bottaro, D.P, Byrd, R.A. | | Deposit date: | 1997-12-18 | | Release date: | 1998-06-24 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal domain of hepatocyte growth factor reveals a potential heparin-binding site.
Structure, 6, 1998
|
|
2JMV
 
 | |
1KBE
 
 | | Solution structure of the cysteine-rich C1 domain of Kinase Suppressor of Ras | | Descriptor: | Kinase Suppressor of Ras, ZINC ION | | Authors: | Zhou, M, Horita, D.A, Waugh, D.S, Byrd, R.A, Morrison, D.K. | | Deposit date: | 2001-11-06 | | Release date: | 2002-01-23 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and functional analysis of the cysteine-rich C1 domain of kinase suppressor of Ras (KSR).
J.Mol.Biol., 315, 2002
|
|
1KBF
 
 | | Solution Structure of the Cysteine-Rich C1 Domain of Kinase Suppressor of Ras | | Descriptor: | Kinase suppressor of Ras 1, ZINC ION | | Authors: | Zhou, M, Horita, D.A, Waugh, D.S, Byrd, R.A, Morrison, D.K. | | Deposit date: | 2001-11-06 | | Release date: | 2002-01-23 | | Last modified: | 2020-02-26 | | Method: | SOLUTION NMR | | Cite: | Solution structure and functional analysis of the cysteine-rich C1 domain of kinase suppressor of Ras (KSR).
J.Mol.Biol., 315, 2002
|
|
2JR0
 
 | | Solution structure of NusB from Aquifex Aeolicus | | Descriptor: | N utilization substance protein B homolog | | Authors: | Das, R, Loss, S, Li, J, Tarasov, S, Wingfield, P, Waugh, D.S, Byrd, R.A, Altieri, A.S. | | Deposit date: | 2007-06-18 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structural biophysics of the NusB:NusE antitermination complex.
J.Mol.Biol., 376, 2008
|
|
2LVN
 
 | | Structure of the gp78 CUE domain | | Descriptor: | E3 ubiquitin-protein ligase AMFR | | Authors: | Liu, S, Chen, Y, Huang, T, Tarasov, S.G, King, A, Li, J, Weissman, A.M, Byrd, R.A, Das, R. | | Deposit date: | 2012-07-09 | | Release date: | 2012-11-21 | | Last modified: | 2012-12-26 | | Method: | SOLUTION NMR | | Cite: | Promiscuous Interactions of gp78 E3 Ligase CUE Domain with Polyubiquitin Chains.
Structure, 20, 2012
|
|
2LVP
 
 | | gp78CUE domain bound to the distal ubiquitin of K48-linked diubiquitin | | Descriptor: | E3 ubiquitin-protein ligase AMFR, Ubiquitin | | Authors: | Liu, S, Chen, Y, Huang, T, Tarasov, S.G, King, A, Li, J, Weissman, A.M, Byrd, R.A, Das, R. | | Deposit date: | 2012-07-09 | | Release date: | 2012-11-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Promiscuous Interactions of gp78 E3 Ligase CUE Domain with Polyubiquitin Chains.
Structure, 20, 2012
|
|
2LVQ
 
 | | gp78CUE domain bound to the proximal ubiquitin of K48-linked diubiquitin | | Descriptor: | E3 ubiquitin-protein ligase AMFR, Ubiquitin | | Authors: | Liu, S, Chen, Y, Huang, T, Tarasov, S.G, King, A, Li, J, Weissman, A.M, Byrd, R.A, Das, R. | | Deposit date: | 2012-07-09 | | Release date: | 2012-11-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Promiscuous Interactions of gp78 E3 Ligase CUE Domain with Polyubiquitin Chains.
Structure, 20, 2012
|
|
2LVO
 
 | | Structure of the gp78CUE domain bound to monubiquitin | | Descriptor: | E3 ubiquitin-protein ligase AMFR, Ubiquitin | | Authors: | Liu, S, Chen, Y, Huang, T, Tarasov, S.G, King, A, Li, J, Weissman, A.M, Byrd, R.A, Das, R. | | Deposit date: | 2012-07-09 | | Release date: | 2012-11-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Promiscuous Interactions of gp78 E3 Ligase CUE Domain with Polyubiquitin Chains.
Structure, 20, 2012
|
|
1DXA
 
 | | BENZO[A]PYRENE DIOL EPOXIDE ADDUCT OF DA IN DUPLEX DNA | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA (5'-D(*CP*TP*CP*GP*GP*GP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*CP*AP*CP*GP*AP*G)-3') | | Authors: | Yeh, H.J.C, Sayer, J.M, Liu, X, Altieri, A.S, Byrd, R.A, Lakshman, M.K, Yagi, H, Schurter, E.J, Gorenstein, D.G, Jerina, D.M. | | Deposit date: | 1995-09-01 | | Release date: | 1995-12-07 | | Last modified: | 2024-03-13 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a nonanucleotide duplex with a dG mismatch opposite a 10S adduct derived from trans addition of a deoxyadenosine N6-amino group to (+)-(7R,8S,9S,10R)-7,8-dihydroxy-9,10-epoxy-7,8,9,10- tetrahydrobenzo[a]pyrene: an unusual syn glycosidic torsion angle at the modified dA
Biochemistry, 34, 1995
|
|
1EY1
 
 | | SOLUTION STRUCTURE OF ESCHERICHIA COLI NUSB | | Descriptor: | ANTITERMINATION FACTOR NUSB | | Authors: | Altieri, A.S, Mazzulla, M.J, Horita, D.A, Coats, R.H, Wingfield, P.T, Byrd, R.A. | | Deposit date: | 2000-05-05 | | Release date: | 2000-06-14 | | Last modified: | 2022-06-15 | | Method: | SOLUTION NMR | | Cite: | The structure of the transcriptional antiterminator NusB from Escherichia coli.
Nat.Struct.Biol., 7, 2000
|
|
1G6Z
 
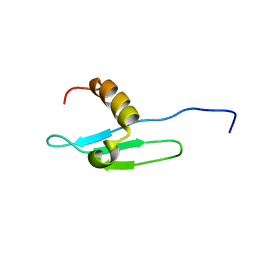 | | SOLUTION STRUCTURE OF THE CLR4 CHROMO DOMAIN | | Descriptor: | CLR4 PROTEIN | | Authors: | Horita, D.A, Ivanova, A.V, Altieri, A.S, Klar, A.J, Byrd, R.A. | | Deposit date: | 2000-11-08 | | Release date: | 2001-04-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure, domain features, and structural implications of mutants of the chromo domain from the fission yeast histone methyltransferase Clr4.
J.Mol.Biol., 307, 2001
|
|
7LEW
 
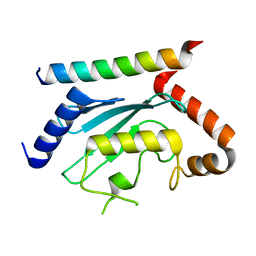 | | Crystal structure of UBE2G2 in complex with the UBE2G2-binding region of AUP1 | | Descriptor: | Lipid droplet-regulating VLDL assembly factor AUP1, Ubiquitin-conjugating enzyme E2 G2 | | Authors: | Liang, Y.-H, Smith, C.E, Tsai, Y.C, Weissman, A.M, Ji, X. | | Deposit date: | 2021-01-15 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.736 Å) | | Cite: | A structurally conserved site in AUP1 binds the E2 enzyme UBE2G2 and is essential for ER-associated degradation.
Plos Biol., 19, 2021
|
|
5C79
 
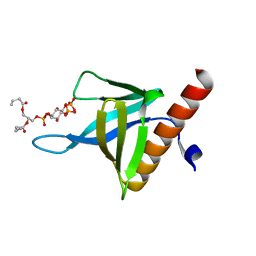 | | PH domain of ASAP1 in complex with diC4-PtdIns(4,5)P2 | | Descriptor: | (2R)-3-{[(R)-HYDROXY{[(1R,2R,3S,4R,5R,6S)-2,3,6-TRIHYDROXY-4,5-BIS(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL]OXY}PROPANE-1 ,2-DIYL DIBUTANOATE, Arf-GAP, CHLORIDE ION | | Authors: | Xia, D, Tang, W.K. | | Deposit date: | 2015-06-24 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular Basis for Cooperative Binding of Anionic Phospholipids to the PH Domain of the Arf GAP ASAP1.
Structure, 23, 2015
|
|
5C6R
 
 | | Crystal structure of PH domain of ASAP1 | | Descriptor: | Arf-GAP, PHOSPHATE ION, TRIETHYLENE GLYCOL | | Authors: | Xia, D, Tang, W.K. | | Deposit date: | 2015-06-23 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis for Cooperative Binding of Anionic Phospholipids to the PH Domain of the Arf GAP ASAP1.
Structure, 23, 2015
|
|
2LXP
 
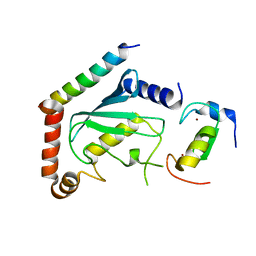 | | NMR structure of two domains in ubiquitin ligase gp78, RING and G2BR, bound to its conjugating enzyme Ube2g | | Descriptor: | E3 ubiquitin-protein ligase AMFR, Ubiquitin-conjugating enzyme E2 G2, ZINC ION | | Authors: | Das, R, Linag, Y, Mariano, J, Li, J, Huang, T, King, A, Weissman, A, Ji, X, Byrd, R. | | Deposit date: | 2012-08-30 | | Release date: | 2013-08-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine.
Embo J., 32, 2013
|
|
2MI8
 
 | |
2LXH
 
 | | NMR structure of the RING domain in ubiquitin ligase gp78 | | Descriptor: | E3 ubiquitin-protein ligase AMFR, ZINC ION | | Authors: | Das, R, Linag, Y, Mariano, J, Li, J, Huang, T, King, A, Weissman, A, Ji, X, Byrd, R. | | Deposit date: | 2012-08-27 | | Release date: | 2013-08-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine.
Embo J., 32, 2013
|
|
4JQU
 
 | | Crystal structure of Ubc7p in complex with the U7BR of Cue1p | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Coupling of ubiquitin conjugation to ER degradation protein 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Liang, Y.-H, Metzger, M.B, Weissman, A.M, Ji, X. | | Deposit date: | 2013-03-20 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A Structurally Unique E2-Binding Domain Activates Ubiquitination by the ERAD E2, Ubc7p, through Multiple Mechanisms.
Mol.Cell, 50, 2013
|
|
