1S17
 
 | | Identification of Novel Potent Bicyclic Peptide Deformylase Inhibitors | | Descriptor: | 2-(3,4-DIHYDRO-3-OXO-2H-BENZO[B][1,4]THIAZIN-2-YL)-N-HYDROXYACETAMIDE, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Molteni, V, He, X, Nabakka, J, Yang, K, Kreusch, A, Gordon, P, Bursulaya, B, Ryder, N.S, Goldberg, R, He, Y. | | Deposit date: | 2004-01-05 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of novel potent bicyclic peptide deformylase inhibitors
Bioorg.Med.Chem.Lett., 14, 2004
|
|
2J14
 
 | | 3,4,5-Trisubstituted Isoxazoles as Novel PPARdelta Agonists: Part2 | | Descriptor: | (3-{4-[2-(2,4-DICHLORO-PHENOXY)-ETHYLCARBAMOYL]-5-PHENYL-ISOXAZOL-3-YL}-PHENYL)-ACETIC ACID, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR DELTA | | Authors: | Epple, R, Azimioara, M, Russo, R, Xie, Y, Wang, X, Cow, C, Wityak, J, Karanewsky, D, Bursulaya, B, Kreusch, A, Tuntland, T, Gerken, A, Iskandar, M, Saez, E, Seidel, H.M, Tian, S.S. | | Deposit date: | 2006-08-08 | | Release date: | 2006-09-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 3,4,5-Trisubstituted Isoxazoles as Novel Ppardelta Agonists. Part 2
Bioorg.Med.Chem.Lett., 16, 2006
|
|
3R5P
 
 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824 | | Descriptor: | Deazaflavin-dependent nitroreductase, SULFATE ION | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-19 | | Release date: | 2012-01-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R5Y
 
 | | Structure of a Deazaflavin-dependent nitroreductase from Nocardia farcinica, with co-factor F420 | | Descriptor: | COENZYME F420, Putative uncharacterized protein | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-20 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R5W
 
 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824, with co-factor F420 | | Descriptor: | COENZYME F420, Deazaflavin-dependent nitroreductase | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-20 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.786 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R5L
 
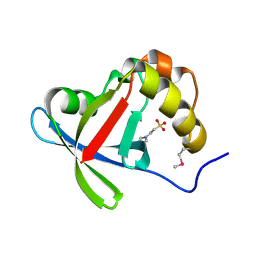 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Deazaflavin-dependent nitroreductase | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayyar, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-18 | | Release date: | 2012-01-18 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R5R
 
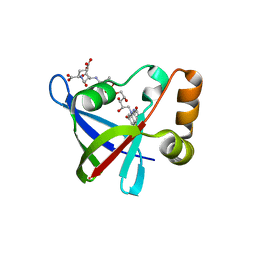 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824, with co-factor F420 | | Descriptor: | COENZYME F420, Deazaflavin-dependent nitroreductase | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-19 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R5Z
 
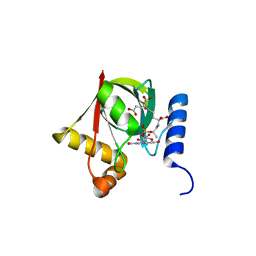 | | Structure of a Deazaflavin-dependent reductase from Nocardia farcinica, with co-factor F420 | | Descriptor: | COENZYME F420, Putative uncharacterized protein, SULFATE ION | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-20 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
4Z55
 
 | |
6UIP
 
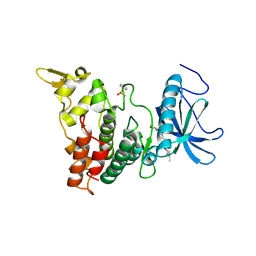 | | DYRK1A Kinase Domain in Complex with a 6-azaindole Derivative, GNF2133. | | Descriptor: | 4-ethyl-N-{4-[1-(oxan-4-yl)-1H-pyrrolo[2,3-c]pyridin-3-yl]pyridin-2-yl}piperazine-1-carboxamide, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | DiDonato, M, Spraggon, G. | | Deposit date: | 2019-10-01 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Selective DYRK1A Inhibitor for the Treatment of Type 1 Diabetes: Discovery of 6-Azaindole Derivative GNF2133.
J.Med.Chem., 63, 2020
|
|
5W5O
 
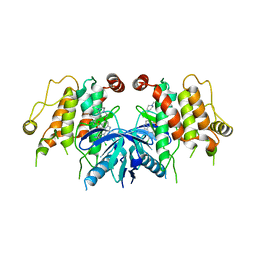 | |
5W5J
 
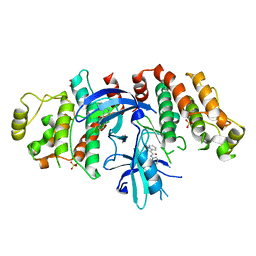 | |
6TCZ
 
 | | Leishmania tarentolae proteasome 20S subunit complexed with LXE408 | | Descriptor: | Proteasome endopeptidase complex, Proteasome subunit alpha type, Proteasome subunit beta, ... | | Authors: | Srinivas, H. | | Deposit date: | 2019-11-07 | | Release date: | 2020-08-26 | | Last modified: | 2020-10-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Discovery and Characterization of Clinical Candidate LXE408 as a Kinetoplastid-Selective Proteasome Inhibitor for the Treatment of Leishmaniases.
J.Med.Chem., 63, 2020
|
|
6TD5
 
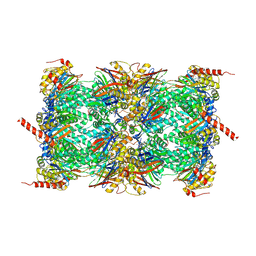 | | Leishmania tarentolae proteasome 20S subunit complexed with LXE408 and bortezomib | | Descriptor: | N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, Proteasome endopeptidase complex, Proteasome subunit alpha type, ... | | Authors: | Srinivas, H. | | Deposit date: | 2019-11-07 | | Release date: | 2020-08-26 | | Last modified: | 2020-10-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Discovery and Characterization of Clinical Candidate LXE408 as a Kinetoplastid-Selective Proteasome Inhibitor for the Treatment of Leishmaniases.
J.Med.Chem., 63, 2020
|
|
7RUN
 
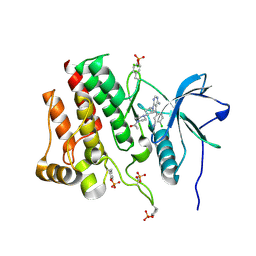 | | Crystal structure of phosphorylated RET tyrosine kinase domain complexed with a pyrrolo[2,3-d]pyrimidine inhibitor. | | Descriptor: | 1-(4-{(1s,3s)-3-[4-amino-5-(3-amino-4-chlorophenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl]cyclobutyl}piperazin-1-yl)ethan-1-one, CHLORIDE ION, Proto-oncogene tyrosine-protein kinase receptor Ret | | Authors: | Lee, C.C, Spraggon, G. | | Deposit date: | 2021-08-17 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Antitarget Selectivity and Tolerability of Novel Pyrrolo[2,3- d ]pyrimidine RET Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
6VHG
 
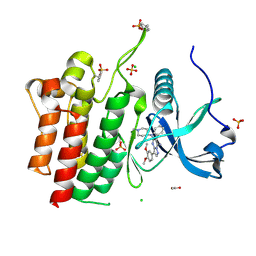 | | Crystal structure of phosphorylated RET tyrosine kinase domain complexed with a pyrazolo[1,5-a]pyrimidine inhibitor | | Descriptor: | 3-(3,4-dimethoxyphenyl)-N~5~-(1-methylpiperidin-4-yl)-6-phenylpyrazolo[1,5-a]pyrimidine-5,7-diamine, ACETATE ION, CHLORIDE ION, ... | | Authors: | Lee, C.C, Spraggon, G. | | Deposit date: | 2020-01-09 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Efficacy and Tolerability of Pyrazolo[1,5-a]pyrimidine RET Kinase Inhibitors for the Treatment of Lung Adenocarcinoma.
Acs Med.Chem.Lett., 11, 2020
|
|
3K5V
 
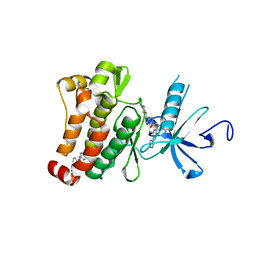 | | Structure of Abl kinase in complex with imatinib and GNF-2 | | Descriptor: | 3-(6-{[4-(trifluoromethoxy)phenyl]amino}pyrimidin-4-yl)benzamide, 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, CHLORIDE ION, ... | | Authors: | Cowan-Jacob, S.W, Fendrich, G, Rummel, G, Strauss, A. | | Deposit date: | 2009-10-08 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Targeting Bcr-Abl by combining allosteric with ATP-binding-site inhibitors.
Nature, 463, 2010
|
|
5FEQ
 
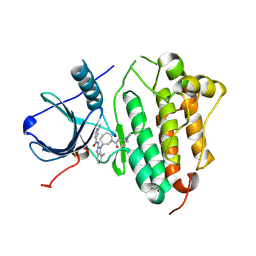 | | EGFR KINASE DOMAIN IN COMPLEX WITH A COVALENT AMINOBENZIMIDAZOLE | | Descriptor: | Epidermal growth factor receptor, ~{N}-[1-[(3~{R})-1-[4-(dimethylamino)but-2-enoyl]azepan-3-yl]-7-methyl-benzimidazol-2-yl]-2-methyl-pyridine-4-carboxamide | | Authors: | DiDonato, M, Spraggon, G. | | Deposit date: | 2015-12-17 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Discovery of (R,E)-N-(7-Chloro-1-(1-[4-(dimethylamino)but-2-enoyl]azepan-3-yl)-1H-benzo[d]imidazol-2-yl)-2-methylisonicotinamide (EGF816), a Novel, Potent, and WT Sparing Covalent Inhibitor of Oncogenic (L858R, ex19del) and Resistant (T790M) EGFR Mutants for the Treatment of EGFR Mutant Non-Small-Cell Lung Cancers.
J.Med.Chem., 59, 2016
|
|
5FED
 
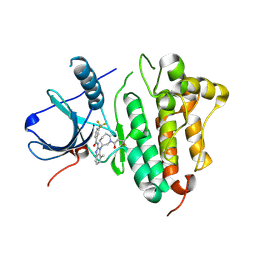 | | EGFR kinase domain in complex with a covalent aminobenzimidazole inhibitor. | | Descriptor: | Epidermal growth factor receptor, ~{N}-[7-methyl-1-[(3~{R})-1-propanoylazepan-3-yl]benzimidazol-2-yl]-3-(trifluoromethyl)benzamide | | Authors: | DiDonato, M, Spraggon, G. | | Deposit date: | 2015-12-16 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Discovery of (R,E)-N-(7-Chloro-1-(1-[4-(dimethylamino)but-2-enoyl]azepan-3-yl)-1H-benzo[d]imidazol-2-yl)-2-methylisonicotinamide (EGF816), a Novel, Potent, and WT Sparing Covalent Inhibitor of Oncogenic (L858R, ex19del) and Resistant (T790M) EGFR Mutants for the Treatment of EGFR Mutant Non-Small-Cell Lung Cancers.
J.Med.Chem., 59, 2016
|
|
5FEE
 
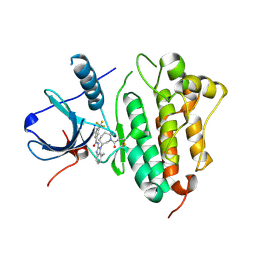 | | EGFR kinase domain T790M mutant in complex with a covalent aminobenzimidazole inhibitor. | | Descriptor: | Epidermal growth factor receptor, ~{N}-[7-methyl-1-[(3~{R})-1-propanoylazepan-3-yl]benzimidazol-2-yl]-3-(trifluoromethyl)benzamide | | Authors: | DiDonato, M, Spraggon, G. | | Deposit date: | 2015-12-16 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of (R,E)-N-(7-Chloro-1-(1-[4-(dimethylamino)but-2-enoyl]azepan-3-yl)-1H-benzo[d]imidazol-2-yl)-2-methylisonicotinamide (EGF816), a Novel, Potent, and WT Sparing Covalent Inhibitor of Oncogenic (L858R, ex19del) and Resistant (T790M) EGFR Mutants for the Treatment of EGFR Mutant Non-Small-Cell Lung Cancers.
J.Med.Chem., 59, 2016
|
|
4AE1
 
 | |
4AE0
 
 | |
6BI2
 
 | |
6BHZ
 
 | | Trastuzumab Fab D185A (Light Chain) Mutant. | | Descriptor: | 1,2-ETHANEDIOL, Trastuzumab Anti-HER2 Fab Heavy Chain, Trastuzumab Anti-HER2 Fab Light Chain D185A | | Authors: | DiDonato, M, Spraggon, G. | | Deposit date: | 2017-10-31 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Tuning a Protein-Labeling Reaction to Achieve Highly Site Selective Lysine Conjugation.
Chembiochem, 19, 2018
|
|
6BI0
 
 | | Trastuzumab Fab N158A, D185A, K190A (Light Chain) Triple Mutant. | | Descriptor: | 1,2-ETHANEDIOL, Trastuzumab anti-HER2 Fab Heavy Chain, Trastuzumab anti-HER2 Fab Light Chain | | Authors: | DiDonato, M, Spraggon, G. | | Deposit date: | 2017-10-31 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.057 Å) | | Cite: | Tuning a Protein-Labeling Reaction to Achieve Highly Site Selective Lysine Conjugation.
Chembiochem, 19, 2018
|
|
