3ZGX
 
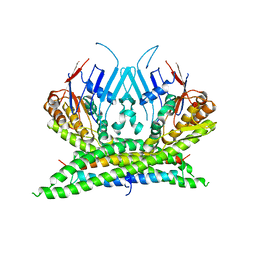 | | Crystal structure of the kleisin-N SMC interface in prokaryotic condensin | | Descriptor: | CHROMOSOME PARTITION PROTEIN SMC, SEGREGATION AND CONDENSATION PROTEIN A | | Authors: | Burmann, F, Shin, H, Basquin, J, Soh, Y, Gimenez, V, Kim, Y, Oh, B, Gruber, S. | | Deposit date: | 2012-12-19 | | Release date: | 2013-01-30 | | Last modified: | 2013-03-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | An Asymmetric Smc-Kleisin Bridge in Prokaryotic Condensin.
Nat.Struct.Mol.Biol., 20, 2013
|
|
6H2X
 
 | | MukB coiled-coil elbow from E. coli | | Descriptor: | Chromosome partition protein MukB,Chromosome partition protein MukB | | Authors: | Buermann, F, Lowe, J. | | Deposit date: | 2018-07-16 | | Release date: | 2019-03-06 | | Last modified: | 2019-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A folded conformation of MukBEF and cohesin.
Nat.Struct.Mol.Biol., 26, 2019
|
|
4I99
 
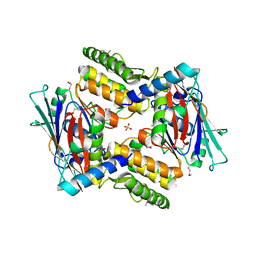 | |
7NYX
 
 | |
7NYW
 
 | | Cryo-EM structure of the MukBEF-MatP-DNA head module | | Descriptor: | 4'-PHOSPHOPANTETHEINE, ADENOSINE-5'-TRIPHOSPHATE, Acyl carrier protein, ... | | Authors: | Buermann, F, Lowe, J. | | Deposit date: | 2021-03-23 | | Release date: | 2021-07-07 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of MukBEF reveals DNA loop entrapment at chromosomal unloading sites.
Mol.Cell, 81, 2021
|
|
7NYY
 
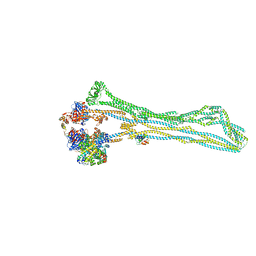 | | Cryo-EM structure of the MukBEF monomer | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, Chromosome partition protein MukB, ... | | Authors: | Buermann, F, Lowe, J. | | Deposit date: | 2021-03-23 | | Release date: | 2021-07-07 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Cryo-EM structure of MukBEF reveals DNA loop entrapment at chromosomal unloading sites.
Mol.Cell, 81, 2021
|
|
7NZ4
 
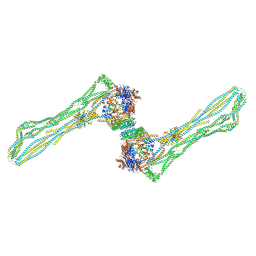 | | Cryo-EM structure of the MukBEF dimer | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, Chromosome partition protein MukB, ... | | Authors: | Buermann, F, Lowe, J. | | Deposit date: | 2021-03-23 | | Release date: | 2021-07-07 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Cryo-EM structure of MukBEF reveals DNA loop entrapment at chromosomal unloading sites.
Mol.Cell, 81, 2021
|
|
7NZ2
 
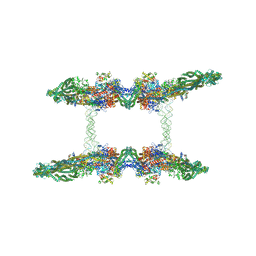 | | Cryo-EM structure of the MukBEF-MatP-DNA tetrad | | Descriptor: | 4'-PHOSPHOPANTETHEINE, ADENOSINE-5'-TRIPHOSPHATE, Acyl carrier protein, ... | | Authors: | Buermann, F, Lowe, J. | | Deposit date: | 2021-03-23 | | Release date: | 2021-07-07 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Cryo-EM structure of MukBEF reveals DNA loop entrapment at chromosomal unloading sites.
Mol.Cell, 81, 2021
|
|
7NYZ
 
 | |
7NZ0
 
 | |
7NZ3
 
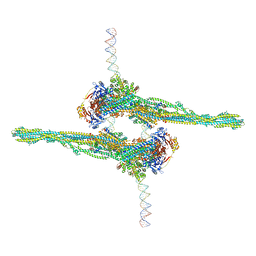 | | Cryo-EM structure of apposed MukBEF-MatP monomers on DNA | | Descriptor: | 4'-PHOSPHOPANTETHEINE, ADENOSINE-5'-TRIPHOSPHATE, Acyl carrier protein, ... | | Authors: | Buermann, F, Lowe, J. | | Deposit date: | 2021-03-23 | | Release date: | 2021-07-07 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Cryo-EM structure of MukBEF reveals DNA loop entrapment at chromosomal unloading sites.
Mol.Cell, 81, 2021
|
|
4I98
 
 | |
4UX3
 
 | | cohesin Smc3-HD:Scc1-N complex from yeast | | Descriptor: | MAGNESIUM ION, MITOTIC CHROMOSOME DETERMINANT-RELATED PROTEIN, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gligoris, T.G, Nasmyth, K, Lowe, J. | | Deposit date: | 2014-08-18 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Closing the Cohesin Ring: Structure and Function of its Smc3-Kleisin Interface.
Science, 346, 2014
|
|
4RSI
 
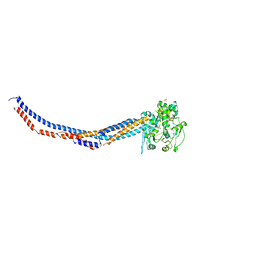 | | Yeast Smc2-Smc4 hinge domain with extended coiled coils | | Descriptor: | PHOSPHATE ION, Structural maintenance of chromosomes protein 2, Structural maintenance of chromosomes protein 4 | | Authors: | Soh, Y.M, Shin, H.C, Oh, B.H. | | Deposit date: | 2014-11-08 | | Release date: | 2014-12-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular Basis for SMC Rod Formation and Its Dissolution upon DNA Binding.
Mol.Cell, 57, 2015
|
|
4RSJ
 
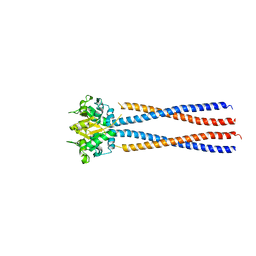 | |
5NNV
 
 | |
5NMO
 
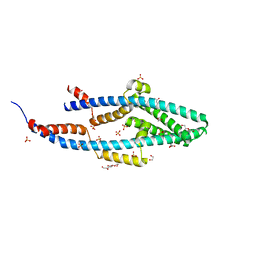 | | Structure of the Bacillus subtilis Smc Joint domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, Chromosome partition protein Smc,Chromosome partition protein Smc, ... | | Authors: | Diebold-Durand, M.-L, Basquin, J, Gruber, S. | | Deposit date: | 2017-04-06 | | Release date: | 2017-06-21 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structure of Full-Length SMC and Rearrangements Required for Chromosome Organization.
Mol. Cell, 67, 2017
|
|
5XEI
 
 | |
5XG3
 
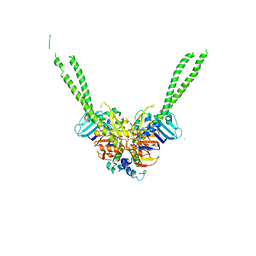 | | Crystal structure of the ATPgS-engaged Smc head domain with an extended coiled coil bound to the C-terminal domain of ScpA derived from Bacillus subtilis | | Descriptor: | COBALT (II) ION, Chromosome partition protein Smc, MAGNESIUM ION, ... | | Authors: | Shin, H.-C, Lee, H, Oh, B.-H. | | Deposit date: | 2017-04-11 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of Full-Length SMC and Rearrangements Required for Chromosome Organization
Mol. Cell, 67, 2017
|
|
5XG2
 
 | |
5XNS
 
 | |
7Q6F
 
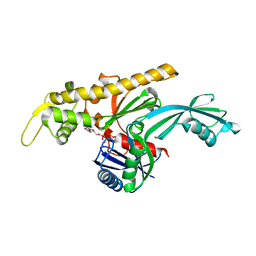 | | Vibrio maritimus FtsA 1-396 ATP, double filament | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division protein FtsA, MAGNESIUM ION | | Authors: | Nierhaus, T, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2021-11-07 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Bacterial divisome protein FtsA forms curved antiparallel double filaments when binding to FtsN.
Nat Microbiol, 7, 2022
|
|
7Q6D
 
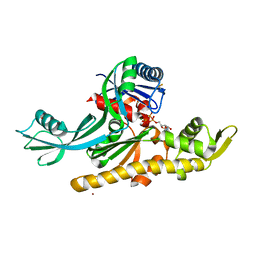 | | E. coli FtsA 1-405 ATP 3 Ni | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division protein FtsA, MAGNESIUM ION, ... | | Authors: | Nierhaus, T, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2021-11-06 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Bacterial divisome protein FtsA forms curved antiparallel double filaments when binding to FtsN.
Nat Microbiol, 7, 2022
|
|
7Q6I
 
 | | Vibrio maritimus FtsA 1-396 ATP and FtsN 1-29, bent tetramers in double filament arrangement | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division protein FtsA, Cell division protein FtsN (polyAla model), ... | | Authors: | Nierhaus, T, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2021-11-07 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Bacterial divisome protein FtsA forms curved antiparallel double filaments when binding to FtsN.
Nat Microbiol, 7, 2022
|
|
7Q6G
 
 | | Xenorhabdus poinarii FtsA 1-396 ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division protein FtsA, MAGNESIUM ION | | Authors: | Nierhaus, T, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2021-11-07 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Bacterial divisome protein FtsA forms curved antiparallel double filaments when binding to FtsN.
Nat Microbiol, 7, 2022
|
|
