4MKR
 
 | | Structure of the apo form of a Zingiber officinale double bond reductase | | Descriptor: | Zingiber officinale double bond reductase | | Authors: | Buratto, J, Langlois d'Estaintot, B, Granier, T, Gallois, B, Willis, M.A, Sang, Y, Flores-Sanchez, I.J, Gang, D.R. | | Deposit date: | 2013-09-05 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure of Zingiber officinale double bond reductase
To be Published
|
|
4LP6
 
 | | Crystal Structure of Human Carbonic Anhydrase II in complex with a quinoline oligoamide foldamer | | Descriptor: | 8-({[4-(3-aminopropoxy)-8-({[4-hydroxy-8-({[4-(2-methylpropoxy)-8-({[4-(3-{[(4-sulfamoylbenzoyl)amino]methyl}phenoxy)butyl]carbamoyl}amino)quinolin-2-yl]carbonyl}amino)quinolin-2-yl]carbonyl}amino)quinolin-2-yl]carbonyl}amino)-4-(carboxymethoxy)quinoline-2-carboxylic acid, Carbonic anhydrase 2, ZINC ION | | Authors: | Buratto, J, Granier, T, Langlois D'estaintot, B, Huc, I, Gallois, B. | | Deposit date: | 2013-07-15 | | Release date: | 2013-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of a complex formed by a protein and a helical aromatic oligoamide foldamer at 2.1 angstrom resolution.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4WE3
 
 | | STRUCTURE OF THE BINARY COMPLEX OF A ZINGIBER OFFICINALE DOUBLE BOND REDUCTASE IN COMPLEX WITH NADP MONOCLINIC CRYSTAL FORM | | Descriptor: | Double Bond Reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Collery, J, Langlois d'Estaintot, B, Buratto, J, Granier, T, Gallois, B, Willis, M.A, Sang, Y, Flores-Sanchez, I.J, Gang, D.R. | | Deposit date: | 2014-09-09 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | STRUCTURE OF ZINGIBER OFFICINALE DOUBLE BOND REDUCTASE
to be published
|
|
4WGG
 
 | | STRUCTURE OF THE TERNARY COMPLEX OF A ZINGIBER OFFICINALE DOUBLE BOND REDUCTASE IN COMPLEX WITH NADP AND CONIFERYL ALDEHYDE | | Descriptor: | (2E)-3-(4-hydroxy-3-methoxyphenyl)prop-2-enal, Double Bond Reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Collery, J, Langlois d'Estaintot, B, Buratto, J, Granier, T, Gallois, B, Willis, M.A, Sang, Y, Flores-Sanchez, I.J, Gang, D.R. | | Deposit date: | 2014-09-18 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | STRUCTURE OF ZINGIBER OFFICINALE DOUBLE BOND REDUCTASE
to be published
|
|
6HFA
 
 | | Crystal structure of hDM2 in complex with a C-terminal triurea capped peptide chimera foldamer. | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, LM266, 1-[(2~{S})-2-azanyl-3-methyl-butyl]urea | | Authors: | Buratto, J, Mauran, L, Goudreau, S, Fribourg, S, Guichard, G. | | Deposit date: | 2018-08-21 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Basis for alpha-Helix Mimicry and Inhibition of Protein-Protein Interactions with Oligourea Foldamers.
Angew.Chem.Int.Ed.Engl., 2020
|
|
6XZJ
 
 | | Structure of zVDR LBD-Calcitriol in complex with chimera 12 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, ACETATE ION, ARG-HIS-LYS-ILE-LEU-URR-UIL-URL-GLN, ... | | Authors: | Buratto, J, Belorusova, A.Y, Rochel, N, Guichard, G. | | Deposit date: | 2020-02-04 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for alpha-Helix Mimicry and Inhibition of Protein-Protein Interactions with Oligourea Foldamers.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6XZH
 
 | | Structure of zVDR LBD-Calcitriol in complex with chimera 10 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, ARG-HIS-LYS-ILE-URL-URK-URL-LEU-GLN, Vitamin D3 receptor A | | Authors: | Buratto, J, Belorusova, A.Y, Rochel, N, Guichard, G. | | Deposit date: | 2020-02-04 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.372 Å) | | Cite: | Structural Basis for alpha-Helix Mimicry and Inhibition of Protein-Protein Interactions with Oligourea Foldamers.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6XZI
 
 | | Structure of zVDR LBD-calcitriol in complex with chimera 11 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, ACETATE ION, ARG-HIS-LYS-ILE-LEU-URK-UIL-URL, ... | | Authors: | Buratto, J, Belorusova, A.Y, Rochel, N, Guichard, G. | | Deposit date: | 2020-02-04 | | Release date: | 2021-02-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for alpha-Helix Mimicry and Inhibition of Protein-Protein Interactions with Oligourea Foldamers.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6XZK
 
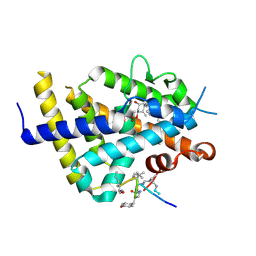 | | Structure of zVDR LBD-Calcitriol in complex with chimera 13 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, ACETATE ION, GLU-ASN-ALA-UIA-URL-URY-URV-UZN-LYS, ... | | Authors: | Buratto, J, Belorusova, A.Y, Rochel, N, Guichard, G. | | Deposit date: | 2020-02-04 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for alpha-Helix Mimicry and Inhibition of Protein-Protein Interactions with Oligourea Foldamers.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6XZV
 
 | | Structure of zVDR LBD-Calcitriol in complex with chimera 18 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, URA-UIA-URL-URY-URV-UZN-LYS, Vitamin D3 receptor A | | Authors: | Buratto, J, Belorusova, A.Y, Rochel, N, Guichard, G. | | Deposit date: | 2020-02-05 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for alpha-Helix Mimicry and Inhibition of Protein-Protein Interactions with Oligourea Foldamers.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
5JWE
 
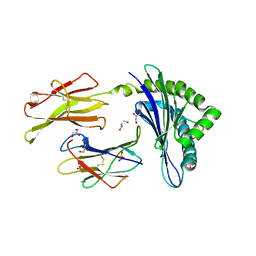 | | Crystal structure of H-2Db in complex with the LCMV-derived GP92-101 peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, H-2 class I histocompatibility antigen, ... | | Authors: | Buratto, J, Badia-Martinez, D, Norstrom, M, Sandalova, T, Achour, A. | | Deposit date: | 2016-05-12 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of H-2Db in complex with the LCMV-derived peptides GP92 and GP392 explain pleiotropic effects of glycosylation on antigen presentation and immunogenicity.
PLoS ONE, 12, 2017
|
|
5JWD
 
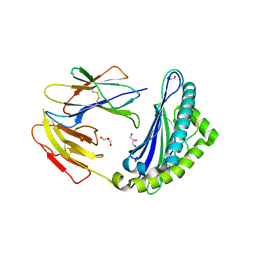 | | Crystal structure of H-2Db in complex with the LCMV-derived GP392-401 peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, H-2 class I histocompatibility antigen, ... | | Authors: | Buratto, J, Badia-Martinez, D, Norstrom, M, Sandalova, T, Achour, A. | | Deposit date: | 2016-05-12 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of H-2Db in complex with the LCMV-derived peptides GP92 and GP392 explain pleiotropic effects of glycosylation on antigen presentation and immunogenicity.
PLoS ONE, 12, 2017
|
|
4NH4
 
 | | Structure of the binary complex of a zingiber officinale double bond reductase in complex with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Zingiber officinale double bond reductase | | Authors: | Langlois D'Estaintot, B, Buratto, J, Granier, T, Gallois, B, Willis, M.A, Sang, Y, Flores-Sanchez, I.J, Gang, D.R. | | Deposit date: | 2013-11-04 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of zingiber officinale double bond reductase
To be Published
|
|
4MTY
 
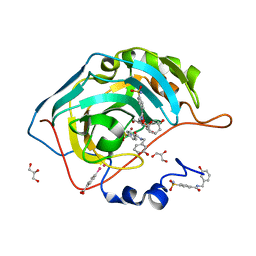 | | Structure at 1A resolution of a helical aromatic foldamer-protein complex. | | Descriptor: | 4-(HYDROXYMERCURY)BENZOIC ACID, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ogayone, T, Buratto, J, Langlois D'Estaintot, B, Stupfel, M, Granier, T, Gallois, B, Huc, Y. | | Deposit date: | 2013-09-20 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure of a complex formed by a protein and a helical aromatic oligoamide foldamer at 2.1 a resolution.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
7PCJ
 
 | | X-ray structure of CypA-C52AK125C/CsA/aromatic foldamer complex | | Descriptor: | 8-azanyl-4-(2-methylpropoxy)quinoline-2-carboxylic acid, 8-azanyl-4-(3-azanylpropoxy)quinoline-2-carboxylic acid, 8-azanyl-4-oxidanyl-quinoline-2-carboxylic acid, ... | | Authors: | Vallade, M, Langlois d'Estaintot, B, Fischer, L, Buratto, J, Savko, M, Huc, I. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | X-ray structure of a cystein mutant of Cyclophilin A tethered to an aromatic oligoamide foldamer complexed with Cyclosporin A
To Be Published
|
|
7QBW
 
 | |
5E8N
 
 | | The structure of the TEIPP associated Trh4 peptide in complex with H-2D(b) | | Descriptor: | Beta-2-microglobulin, Ceramide synthase 5, GLYCEROL, ... | | Authors: | Hafstrand, I, Doorduijn, E, Duru, A.D, Buratto, J, Oliveira, C.C, Sandalova, T, van Hall, T, Achour, A. | | Deposit date: | 2015-10-14 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The MHC Class I Cancer-Associated Neoepitope Trh4 Linked with Impaired Peptide Processing Induces a Unique Noncanonical TCR Conformer.
J Immunol., 196, 2016
|
|
5E8O
 
 | | The structure of the TEIPP associated altered peptide ligand Trh4-p2ABU in complex with H-2D(b) | | Descriptor: | Beta-2-microglobulin, Ceramide synthase 5, H-2 class I histocompatibility antigen, ... | | Authors: | Hafstrand, I, Doorduijn, E, Duru, A.D, Buratto, J, Oliveira, C.C, Sandalova, T, van Hall, T, Achour, A. | | Deposit date: | 2015-10-14 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The MHC Class I Cancer-Associated Neoepitope Trh4 Linked with Impaired Peptide Processing Induces a Unique Noncanonical TCR Conformer.
J Immunol., 196, 2016
|
|
5E8P
 
 | | The structure of the TEIPP associated altered peptide ligand Trh4-p5NLE in complex with H-2D(b) | | Descriptor: | Beta-2-microglobulin, Ceramide synthase 5, H-2 class I histocompatibility antigen, ... | | Authors: | Hafstrand, I, Doorduijn, E, Duru, A.D, Buratto, J, Oliveira, C.C, Sandalova, T, van Hall, T, Achour, A. | | Deposit date: | 2015-10-14 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The MHC Class I Cancer-Associated Neoepitope Trh4 Linked with Impaired Peptide Processing Induces a Unique Noncanonical TCR Conformer.
J Immunol., 196, 2016
|
|
6I8C
 
 | | Crystal structure of the murine beta-2-microglobulin. | | Descriptor: | Beta-2-microglobulin, TRIETHYLENE GLYCOL | | Authors: | Achour, A, Sandalova, T, Ricagno, S, Sun, R. | | Deposit date: | 2018-11-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Biochemical and biophysical comparison of human and mouse beta-2 microglobulin reveals the molecular determinants of low amyloid propensity.
Febs J., 287, 2020
|
|
