1W2W
 
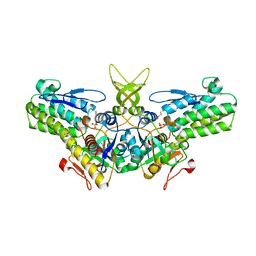 | | Crystal structure of yeast Ypr118w, a methylthioribose-1-phosphate isomerase related to regulatory eIF2B subunits | | Descriptor: | 5-METHYLTHIORIBOSE-1-PHOSPHATE ISOMERASE, SULFATE ION | | Authors: | Bumann, M, Djafarzadeh, S, Oberholzer, A.E, Bigler, P, Altmann, M, Trachsel, H, Baumann, U. | | Deposit date: | 2004-07-09 | | Release date: | 2004-07-16 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Yeast Ypr118W, a Methylthioribose-1-Phosphate Isomerase Related to Regulatory Eif2B Subunits
J.Biol.Chem., 279, 2004
|
|
5I6A
 
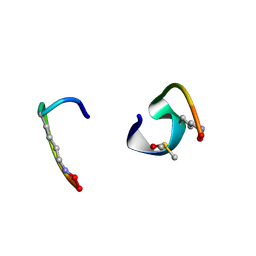 | | bicyclo[3.3.2]decapeptide | | Descriptor: | ALA-PHE-GLY-LYD-VAL-PHE-PRO-GLN-ALA-GLY, DIMETHYL SULFOXIDE | | Authors: | Bartoloni, M, Waltersperger, S, Bumann, M, Stocker, A, Darbre, T, Reymond, J.-L. | | Deposit date: | 2016-02-16 | | Release date: | 2016-03-09 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (0.813 Å) | | Cite: | Stereoselective synthesis and structure determination of a bicyclo[3.3.2]decapeptide
Arkivoc, 2014, 2014
|
|
3HBV
 
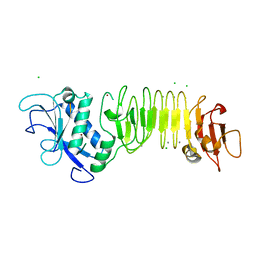 | | PrtC methionine mutants: M226A in-house | | Descriptor: | CALCIUM ION, CHLORIDE ION, Secreted protease C, ... | | Authors: | Oberholzer, A.E, Bumann, M, Hege, T, Russo, S, Baumann, U. | | Deposit date: | 2009-05-05 | | Release date: | 2009-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Metzincin's canonical methionine is responsible for the structural integrity of the zinc-binding site
Biol.Chem., 390, 2009
|
|
4GMK
 
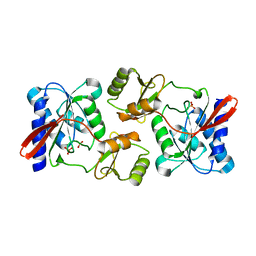 | | Crystal Structure of Ribose 5-Phosphate Isomerase from the Probiotic Bacterium Lactobacillus salivarius UCC118 | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, Ribose-5-phosphate isomerase A | | Authors: | Lobley, C.M.C, Aller, P, Douangamath, A, Reddivari, Y, Bumann, M, Bird, L.E, Brandao-Neto, J, Owens, R.J, O'Toole, P.W, Walsh, M.A. | | Deposit date: | 2012-08-16 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure of ribose 5-phosphate isomerase from the probiotic bacterium Lactobacillus salivarius UCC118.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
2VSX
 
 | | Crystal Structure of a Translation Initiation Complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP-DEPENDENT RNA HELICASE EIF4A, EUKARYOTIC INITIATION FACTOR 4F SUBUNIT P150 | | Authors: | Schutz, P, Bumann, M, Oberholzer, A.E, Bieniossek, C, Altmann, M, Trachsel, H, Baumann, U. | | Deposit date: | 2008-04-30 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Yeast Eif4A-Eif4G Complex: An RNA-Helicase Controlled by Protein-Protein Interactions.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2VSO
 
 | | Crystal Structure of a Translation Initiation Complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP-DEPENDENT RNA HELICASE EIF4A, EUKARYOTIC INITIATION FACTOR 4F SUBUNIT P150 | | Authors: | Schutz, P, Bumann, M, Oberholzer, A.E, Bieniossek, C, Altmann, M, Trachsel, H, Baumann, U. | | Deposit date: | 2008-04-28 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Yeast Eif4A-Eif4G Complex: An RNA-Helicase Controlled by Protein-Protein Interactions.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
4C7V
 
 | | Apo Transketolase from Lactobacillus salivarius at 2.2A resolution | | Descriptor: | TRANSKETOLASE | | Authors: | Lobley, C.M.C, Lukacik, P, Bumann, M, Aller, P, Douangamath, A, O'Toole, P.W, Walsh, M.A. | | Deposit date: | 2013-09-26 | | Release date: | 2014-10-08 | | Last modified: | 2015-10-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High Resolution Crystal Structures of Lactobacillus Salivarius Transketolase in the Presence and Absence of Thiamine Pyrophosphate
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4C7X
 
 | | Thiamine Pyrophosphate Bound Transketolase from Lactobacillus salivarius at 2.2A resolution | | Descriptor: | MAGNESIUM ION, THIAMINE DIPHOSPHATE, TRANSKETOLASE | | Authors: | Lobley, C.M.C, Lukacik, P, Bumann, M, Aller, P, Douangamath, A, O'Toole, P.W, Walsh, M.A. | | Deposit date: | 2013-09-26 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | High Resolution Crystal Structures of Lactobacillus Salivarius Transketolase in the Presence and Absence of Thiamine Pyrophosphate
Acta Crystallogr.,Sect.F, 71, 2015
|
|
3HBU
 
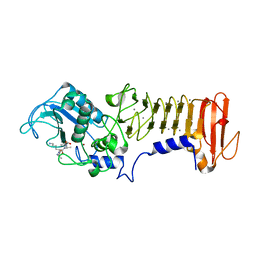 | | PrtC methionine mutants: M226H DESY | | Descriptor: | CALCIUM ION, CHLORIDE ION, Secreted protease C, ... | | Authors: | Oberholzer, A.E, Bumann, M, Hege, T, Russo, S, Baumann, U. | | Deposit date: | 2009-05-05 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Metzincin's canonical methionine is responsible for the structural integrity of the zinc-binding site
Biol.Chem., 390, 2009
|
|
3HB2
 
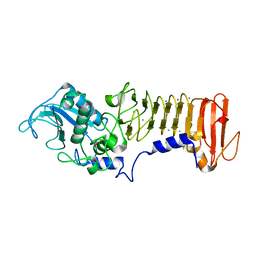 | | PrtC methionine mutants: M226I | | Descriptor: | CALCIUM ION, CHLORIDE ION, Secreted protease C, ... | | Authors: | Oberholzer, A.E, Bumann, M, Hege, T, Russo, S, Baumann, U. | | Deposit date: | 2009-05-04 | | Release date: | 2009-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Metzincin's canonical methionine is responsible for the structural integrity of the zinc-binding site
Biol.Chem., 390, 2009
|
|
3HDA
 
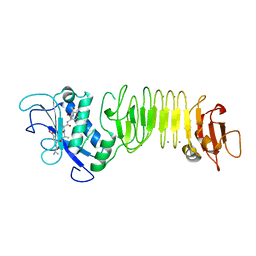 | | PrtC methionine mutants: M226A_DESY | | Descriptor: | CALCIUM ION, CHLORIDE ION, Secreted protease C, ... | | Authors: | Oberholzer, A.E, Bumann, M, Hege, T, Russo, S, Baumann, U. | | Deposit date: | 2009-05-07 | | Release date: | 2009-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.131 Å) | | Cite: | Metzincin's canonical methionine is responsible for the structural integrity of the zinc-binding site
Biol.Chem., 390, 2009
|
|
2BG5
 
 | | Crystal Structure of the Phosphoenolpyruvate-binding Enzyme I-Domain from the Thermoanaerobacter tengcongensis PEP: Sugar Phosphotransferase System (PTS) | | Descriptor: | PHOSPHOENOLPYRUVATE-PROTEIN KINASE | | Authors: | Oberholzer, A.E, Bumann, M, Schneider, P, Baechler, C, Siebold, C, Baumann, U, Erni, B. | | Deposit date: | 2004-12-17 | | Release date: | 2005-02-02 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal Structure of the Phosphoenolpyruvate-Binding Enzyme I-Domain from the Thermoanaerobacter Tengcongensis Pep: Sugar Phosphotransferase System (Pts)
J.Mol.Biol., 346, 2005
|
|
2IU1
 
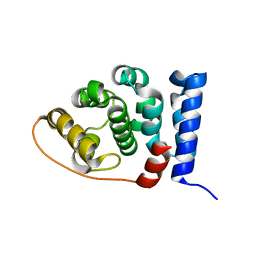 | |
2CE7
 
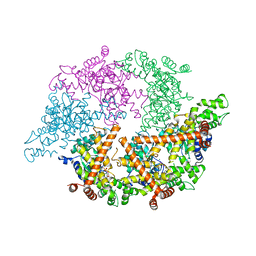 | | EDTA treated | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CELL DIVISION PROTEIN FTSH, MAGNESIUM ION, ... | | Authors: | Bieniossek, C, Baumann, U. | | Deposit date: | 2006-02-03 | | Release date: | 2006-02-09 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | The Molecular Architecture of the Metalloprotease Ftsh.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2CEA
 
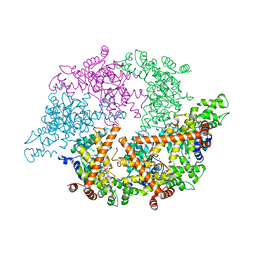 | | CELL DIVISION PROTEIN FTSH | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CELL DIVISION PROTEIN FTSH, MAGNESIUM ION, ... | | Authors: | Bieniossek, C, Baumann, U. | | Deposit date: | 2006-02-03 | | Release date: | 2006-02-09 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Molecular Architecture of the Metalloprotease Ftsh.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
4MDZ
 
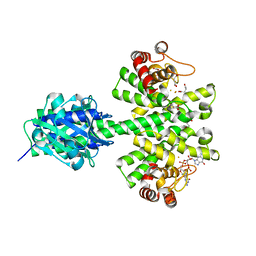 | | Crystal structure of a HD-GYP domain (a cyclic-di-GMP phosphodiesterase) containing a tri-nuclear metal centre | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), FE (III) ION, Metal dependent phosphohydrolase, ... | | Authors: | Bellini, D, Walsh, M.A, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2013-08-23 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal structure of an HD-GYP domain cyclic-di-GMP phosphodiesterase reveals an enzyme with a novel trinuclear catalytic iron centre.
Mol.Microbiol., 91, 2014
|
|
4ME4
 
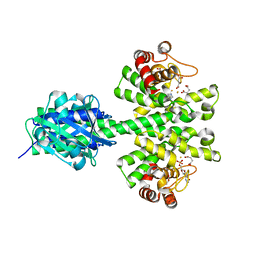 | |
4MCW
 
 | | Crystal structure of a HD-GYP domain (a cyclic-di-GMP phosphodiesterase) containing a tri-nuclear metal centre | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, IMIDAZOLE, ... | | Authors: | Bellini, D, Walsh, M.A, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2013-08-21 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of an HD-GYP domain cyclic-di-GMP phosphodiesterase reveals an enzyme with a novel trinuclear catalytic iron centre.
Mol.Microbiol., 91, 2014
|
|
