1B1A
 
 | | GLUTAMATE MUTASE (B12-BINDING SUBUNIT), NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | GLUTAMATE MUTASE | | Authors: | Hoffmann, B, Konrat, R, Bothe, H, Buckel, W, Kraeutler, B. | | Deposit date: | 1998-11-19 | | Release date: | 1999-07-13 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of the B12-binding subunit of glutamate mutase from Clostridium cochlearium.
Eur.J.Biochem., 263, 1999
|
|
7ZC6
 
 | | Na+ - translocating ferredoxin: NAD+ reductase (Rnf) of C. tetanomorphum | | Descriptor: | FE (III) ION, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Ermler, U, Vitt, S, Buckel, W. | | Deposit date: | 2022-03-25 | | Release date: | 2022-09-28 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Purification and structural characterization of the Na + -translocating ferredoxin: NAD + reductase (Rnf) complex of Clostridium tetanomorphum.
Nat Commun, 13, 2022
|
|
5OL2
 
 | | The electron transferring flavoprotein/butyryl-CoA dehydrogenase complex from Clostridium difficile | | Descriptor: | Acyl-CoA dehydrogenase, CALCIUM ION, COENZYME A PERSULFIDE, ... | | Authors: | Demmer, J.K, Chowdhury, N.P, Selmer, T, Ermler, U, Buckel, W. | | Deposit date: | 2017-07-26 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The semiquinone swing in the bifurcating electron transferring flavoprotein/butyryl-CoA dehydrogenase complex from Clostridium difficile.
Nat Commun, 8, 2017
|
|
4KPU
 
 | | Electron transferring flavoprotein of Acidaminococcus fermentans: Towards a mechanism of flavin-based electron bifurcation | | Descriptor: | CHLORIDE ION, Electron transfer flavoprotein alpha subunit, Electron transfer flavoprotein alpha/beta-subunit, ... | | Authors: | Mowafy, A.M, Chowdhury, N.P, Demmer, J, Upadhyay, V, Kolzer, S, Jayamani, E, Kahnt, J, Demmer, U, Ermler, U, Buckel, W. | | Deposit date: | 2013-05-14 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Studies on the Mechanism of Electron Bifurcation Catalyzed by Electron Transferring Flavoprotein (Etf) and Butyryl-CoA Dehydrogenase (Bcd) of Acidaminococcus fermentans.
J.Biol.Chem., 289, 2014
|
|
1U8V
 
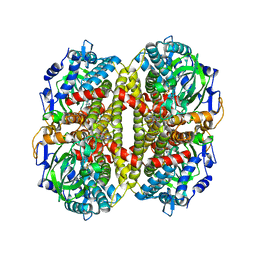 | | Crystal Structure of 4-Hydroxybutyryl-CoA Dehydratase from Clostridium aminobutyricum: Radical catalysis involving a [4Fe-4S] cluster and flavin | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Gamma-aminobutyrate metabolism dehydratase/isomerase, IRON/SULFUR CLUSTER | | Authors: | Martins, B.M, Dobbek, H, Cinkaya, I, Buckel, W, Messerschmidt, A. | | Deposit date: | 2004-08-07 | | Release date: | 2004-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of 4-hydroxybutyryl-CoA dehydratase: radical catalysis involving a [4Fe-4S] cluster and flavin.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1XDW
 
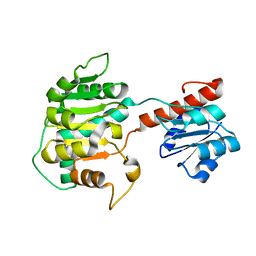 | | NAD+-dependent (R)-2-Hydroxyglutarate Dehydrogenase from Acidaminococcus fermentans | | Descriptor: | NAD+-dependent (R)-2-Hydroxyglutarate Dehydrogenase | | Authors: | Martins, B.M, Macedo-Ribeiro, S, Bresser, J, Buckel, W, Messerschmidt, A. | | Deposit date: | 2004-09-08 | | Release date: | 2005-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for stereo-specific catalysis in NAD(+)-dependent (R)-2-hydroxyglutarate dehydrogenase from Acidaminococcus fermentans.
Febs J., 272, 2005
|
|
1PIX
 
 | | Crystal structure of the carboxyltransferase subunit of the bacterial ion pump glutaconyl-coenzyme A decarboxylase | | Descriptor: | FORMIC ACID, Glutaconyl-CoA decarboxylase A subunit, SULFATE ION | | Authors: | Wendt, K.S, Schall, I, Huber, R, Buckel, W, Jacob, U. | | Deposit date: | 2003-05-30 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the carboxyltransferase subunit of the bacterial sodium ion pump glutaconyl-coenzyme A decarboxylase
Embo J., 22, 2003
|
|
1POI
 
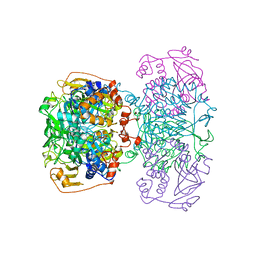 | | CRYSTAL STRUCTURE OF GLUTACONATE COENZYME A-TRANSFERASE FROM ACIDAMINOCOCCUS FERMENTANS TO 2.55 ANGSTOMS RESOLUTION | | Descriptor: | COPPER (II) ION, GLUTACONATE COENZYME A-TRANSFERASE | | Authors: | Jacob, U, Mack, M, Clausen, T, Huber, R, Buckel, W, Messerschmidt, A. | | Deposit date: | 1997-01-24 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Glutaconate CoA-transferase from Acidaminococcus fermentans: the crystal structure reveals homology with other CoA-transferases.
Structure, 5, 1997
|
|
1HUX
 
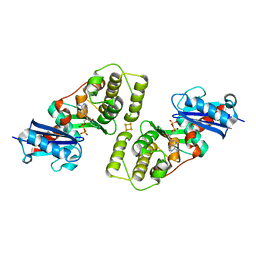 | | CRYSTAL STRUCTURE OF THE ACIDAMINOCOCCUS FERMENTANS (R)-2-HYDROXYGLUTARYL-COA DEHYDRATASE COMPONENT A | | Descriptor: | ACTIVATOR OF (R)-2-HYDROXYGLUTARYL-COA DEHYDRATASE, ADENOSINE-5'-DIPHOSPHATE, IRON/SULFUR CLUSTER | | Authors: | Locher, K.P, Hans, M, Yeh, A.P, Schmid, B, Buckel, W, Rees, D.C. | | Deposit date: | 2001-01-04 | | Release date: | 2001-03-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the Acidaminococcus fermentans 2-hydroxyglutaryl-CoA dehydratase component A.
J.Mol.Biol., 307, 2001
|
|
3O3N
 
 | | (R)-2-hydroxyisocaproyl-CoA dehydratase in complex with its substrate (R)-2-hydroxyisocaproyl-CoA | | Descriptor: | HYDROSULFURIC ACID, IRON/SULFUR CLUSTER, S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-3-phosphonooxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl ]oxy-hydroxy-phosphoryl]oxy-2-hydroxy-3,3-dimethyl-butanoyl]amino]propanoylamino]ethyl] (2R)-2-hydroxy-4-methyl-pentanethioate, ... | | Authors: | Knauer, S.H, Buckel, W, Dobbek, H. | | Deposit date: | 2010-07-25 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Reductive Radical Formation and Electron Recycling in (R)-2-Hydroxyisocaproyl-CoA Dehydratase.
J.Am.Chem.Soc., 133, 2011
|
|
3O3M
 
 | | (R)-2-Hydroxyisocaproyl-CoA Dehydratase | | Descriptor: | HYDROSULFURIC ACID, IRON/SULFUR CLUSTER, SULFATE ION, ... | | Authors: | Knauer, S.H, Buckel, W, Dobbek, H. | | Deposit date: | 2010-07-25 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Basis for Reductive Radical Formation and Electron Recycling in (R)-2-Hydroxyisocaproyl-CoA Dehydratase.
J.Am.Chem.Soc., 133, 2011
|
|
3O3O
 
 | | (R)-2-hydroxyisocaproyl-CoA dehydratase in complex with (R)-2-hydroxyisocaproate | | Descriptor: | (2R)-2-hydroxy-4-methylpentanoic acid, HYDROSULFURIC ACID, IRON/SULFUR CLUSTER, ... | | Authors: | Knauer, S.H, Buckel, W, Dobbek, H. | | Deposit date: | 2010-07-25 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Reductive Radical Formation and Electron Recycling in (R)-2-Hydroxyisocaproyl-CoA Dehydratase.
J.Am.Chem.Soc., 133, 2011
|
|
4L2I
 
 | | Electron transferring flavoprotein of Acidaminococcus fermentans: Towards a mechanism of flavin-based electron bifurcation | | Descriptor: | CHLORIDE ION, Electron transfer flavoprotein alpha subunit, Electron transfer flavoprotein alpha/beta-subunit, ... | | Authors: | Mowafy, A.M, Chowdhury, N.P, Demmer, J, Upadhyay, V, Kolzer, S, Jayamani, E, Kahnt, J, Demmer, U, Ermler, U, Buckel, W. | | Deposit date: | 2013-06-04 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Studies on the Mechanism of Electron Bifurcation Catalyzed by Electron Transferring Flavoprotein (Etf) and Butyryl-CoA Dehydrogenase (Bcd) of Acidaminococcus fermentans.
J.Biol.Chem., 289, 2014
|
|
4L1F
 
 | | Electron transferring flavoprotein of Acidaminococcus fermentans: Towards a mechanism of flavin-based electron bifurcation | | Descriptor: | 1,3-PROPANDIOL, Acyl-CoA dehydrogenase domain protein, COENZYME A PERSULFIDE, ... | | Authors: | Mowafy, A.M, Chowdhury, N.P, Demmer, J, Upadhyay, V, Kolzer, S, Jayamani, E, Kahnt, J, Demmer, U, Ermler, U, Buckel, W. | | Deposit date: | 2013-06-03 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Studies on the Mechanism of Electron Bifurcation Catalyzed by Electron Transferring Flavoprotein (Etf) and Butyryl-CoA Dehydrogenase (Bcd) of Acidaminococcus fermentans.
J.Biol.Chem., 289, 2014
|
|
3GF7
 
 | | Glutaconyl-coA decarboxylase A subunit from Clostridium symbiosum apoprotein | | Descriptor: | Glutaconyl-CoA decarboxylase subunit A, SULFATE ION | | Authors: | Kress, D, Brugel, D, Buckel, W, Essen, L.-O. | | Deposit date: | 2009-02-26 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An asymmetric model for Na+-translocating glutaconyl-CoA decarboxylases
J.Biol.Chem., 284, 2009
|
|
3GF3
 
 | | Glutaconyl-coA decarboxylase A subunit from Clostridium symbiosum co-crystallized with glutaconyl-coA | | Descriptor: | CHLORIDE ION, CROTONYL COENZYME A, Glutaconyl-CoA decarboxylase subunit A | | Authors: | Kress, D, Brugel, D, Buckel, W, Essen, L.-O. | | Deposit date: | 2009-02-26 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An asymmetric model for Na+-translocating glutaconyl-CoA decarboxylases
J.Biol.Chem., 284, 2009
|
|
3GMA
 
 | | Glutaconyl-coA decarboxylase A subunit from Clostridium symbiosum co-crystallized with glutaryl-CoA | | Descriptor: | Glutaconyl-CoA decarboxylase subunit A, glutaryl-coenzyme A | | Authors: | Kress, D, Brugel, D, Buckel, W, Essen, L.-O. | | Deposit date: | 2009-03-13 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An asymmetric model for Na+-translocating glutaconyl-CoA decarboxylases
J.Biol.Chem., 284, 2009
|
|
3GLM
 
 | | Glutaconyl-coA decarboxylase A subunit from Clostridium symbiosum co-crystallized with crotonyl-coA | | Descriptor: | CHLORIDE ION, CROTONYL COENZYME A, Glutaconyl-CoA decarboxylase subunit A | | Authors: | Kress, D, Brugel, D, Buckel, W, Essen, L.-O. | | Deposit date: | 2009-03-12 | | Release date: | 2009-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An asymmetric model for Na+-translocating glutaconyl-CoA decarboxylases
J.Biol.Chem., 284, 2009
|
|
1CB7
 
 | |
1CCW
 
 | |
1I9C
 
 | |
5OW0
 
 | | Crystal structure of an electron transfer flavoprotein from Geobacter metallireducens | | Descriptor: | ADENOSINE MONOPHOSPHATE, Electron transfer flavoprotein, alpha subunit, ... | | Authors: | Essen, L.-O, Vogt, M.S, Heider, J, Koelzer, S, Peschke, P, Chowdhury, N.P, Schuehle, K, Kleinsorge, D. | | Deposit date: | 2017-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2019-09-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional characterization of an electron transfer flavoprotein involved in toluene degradation in strictly anaerobic bacteria.
J.Bacteriol., 2019
|
|
2Y8N
 
 | | Crystal structure of glycyl radical enzyme | | Descriptor: | 4-HYDROXYPHENYLACETATE DECARBOXYLASE LARGE SUBUNIT, 4-HYDROXYPHENYLACETATE DECARBOXYLASE SMALL SUBUNIT, IRON/SULFUR CLUSTER | | Authors: | Martins, B.M, Blaser, M, Feliks, M, Ullmann, G.M, Selmer, T. | | Deposit date: | 2011-02-08 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for a Kolbe-Type Decarboxylation Catalyzed by a Glycyl Radical Enzyme.
J.Am.Chem.Soc., 133, 2011
|
|
2YAJ
 
 | | CRYSTAL STRUCTURE OF GLYCYL RADICAL ENZYME with bound substrate | | Descriptor: | 4-HYDROXYPHENYLACETATE, 4-HYDROXYPHENYLACETATE DECARBOXYLASE LARGE SUBUNIT, 4-HYDROXYPHENYLACETATE DECARBOXYLASE SMALL SUBUNIT, ... | | Authors: | Martins, B.M, Blaser, M, Feliks, M, Ullmann, G.M, Selmer, T. | | Deposit date: | 2011-02-23 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.813 Å) | | Cite: | Structural Basis for a Kolbe-Type Decarboxylation Catalyzed by a Glycyl Radical Enzyme.
J.Am.Chem.Soc., 133, 2011
|
|
1MUM
 
 | |
