1MRP
 
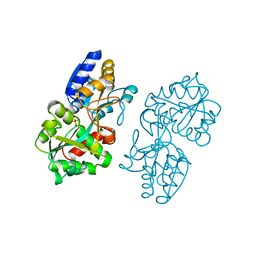 | | FERRIC-BINDING PROTEIN FROM HAEMOPHILUS INFLUENZAE | | Descriptor: | FE (III) ION, FERRIC IRON BINDING PROTEIN, PHOSPHATE ION | | Authors: | Bruns, C.M, Nowalk, A.J, Arvai, A.S, Mctigue, M.A, Vaughan, K.G, Mietzner, T.A, Mcree, D.E. | | Deposit date: | 1997-05-14 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Haemophilus influenzae Fe(+3)-binding protein reveals convergent evolution within a superfamily.
Nat.Struct.Biol., 4, 1997
|
|
1FNC
 
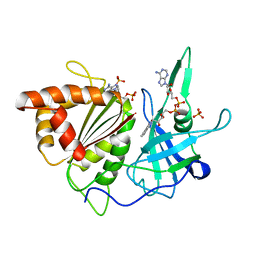 | | REFINED CRYSTAL STRUCTURE OF SPINACH FERREDOXIN REDUCTASE AT 1.7 ANGSTROMS RESOLUTION: OXIDIZED, REDUCED, AND 2'-PHOSPHO-5'-AMP BOUND STATES | | Descriptor: | ADENOSINE-2'-5'-DIPHOSPHATE, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, FERREDOXIN-NADP+ REDUCTASE, ... | | Authors: | Bruns, C.M, Karplus, P.A. | | Deposit date: | 1995-01-05 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined crystal structure of spinach ferredoxin reductase at 1.7 A resolution: oxidized, reduced and 2'-phospho-5'-AMP bound states.
J.Mol.Biol., 247, 1995
|
|
1FNB
 
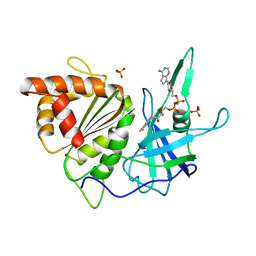 | | REFINED CRYSTAL STRUCTURE OF SPINACH FERREDOXIN REDUCTASE AT 1.7 ANGSTROMS RESOLUTION: OXIDIZED, REDUCED, AND 2'-PHOSPHO-5'-AMP BOUND STATES | | Descriptor: | FERREDOXIN-NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, ... | | Authors: | Bruns, C.M, Karplus, P.A. | | Deposit date: | 1995-01-05 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Refined crystal structure of spinach ferredoxin reductase at 1.7 A resolution: oxidized, reduced and 2'-phospho-5'-AMP bound states.
J.Mol.Biol., 247, 1995
|
|
1FND
 
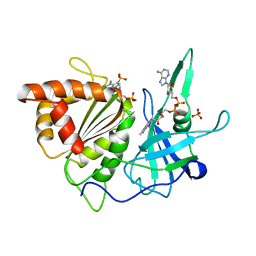 | | REFINED CRYSTAL STRUCTURE OF SPINACH FERREDOXIN REDUCTASE AT 1.7 ANGSTROMS RESOLUTION: OXIDIZED, REDUCED, AND 2'-PHOSPHO-5'-AMP BOUND STATES | | Descriptor: | ADENOSINE-2'-5'-DIPHOSPHATE, FERREDOXIN-NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Bruns, C.M, Karplus, P.A. | | Deposit date: | 1995-01-05 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Refined crystal structure of spinach ferredoxin reductase at 1.7 A resolution: oxidized, reduced and 2'-phospho-5'-AMP bound states.
J.Mol.Biol., 247, 1995
|
|
1GUM
 
 | | HUMAN GLUTATHIONE TRANSFERASE A4-4 WITHOUT LIGANDS | | Descriptor: | PROTEIN (GLUTATHIONE TRANSFERASE A4-4) | | Authors: | Bruns, C.M, Hubatsch, I, Ridderstrom, M, Mannervik, B, Tainer, J.A. | | Deposit date: | 1998-06-11 | | Release date: | 1999-01-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human glutathione transferase A4-4 crystal structures and mutagenesis reveal the basis of high catalytic efficiency with toxic lipid peroxidation products
J.Mol.Biol., 288, 1999
|
|
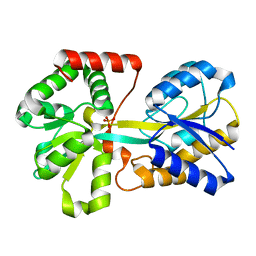
1GUL
 
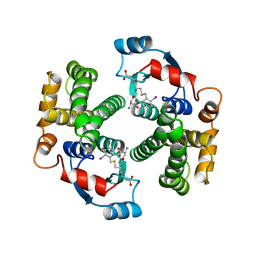 | | HUMAN GLUTATHIONE TRANSFERASE A4-4 COMPLEX WITH IODOBENZYL GLUTATHIONE | | Descriptor: | GAMMA-GLUTAMYL[S-(2-IODOBENZYL)CYSTEINYL]GLYCINE, Glutathione S-transferase A4 | | Authors: | Bruns, C.M, Hubatsch, I, Ridderstrom, M, Mannervik, B, Tainer, J.A. | | Deposit date: | 1998-06-10 | | Release date: | 1999-01-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human glutathione transferase A4-4 crystal structures and mutagenesis reveal the basis of high catalytic efficiency with toxic lipid peroxidation products
J.Mol.Biol., 288, 1999
|
|
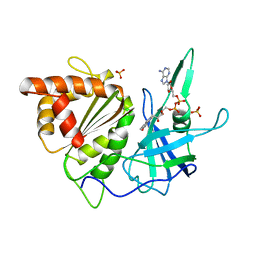
1D9V
 
 | |
1FRN
 
 | |
1D9Y
 
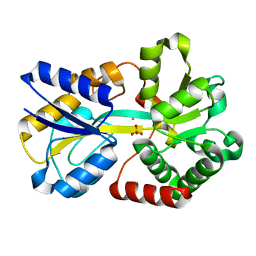 | | NEISSERIA GONORRHOEAE FERRIC BINDING PROTEIN | | Descriptor: | FE (III) ION, PHOSPHATE ION, PROTEIN (PERIPLASMIC IRON-BINDING PROTEIN) | | Authors: | McRee, D.E, Bruns, C.M, Williams, P.A, Mietzner, T.A, Nunn, R. | | Deposit date: | 1999-10-30 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Iron Uptake in the Pathogen Neisseria gonorrhoeae
To be Published
|
|
1TLL
 
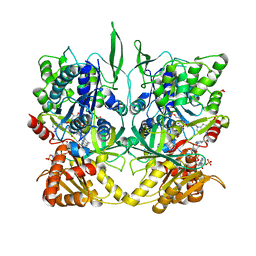 | | CRYSTAL STRUCTURE OF RAT NEURONAL NITRIC-OXIDE SYNTHASE REDUCTASE MODULE AT 2.3 A RESOLUTION. | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Garcin, E.D, Bruns, C.M, Lloyd, S.J, Hosfield, D.J, Tiso, M, Gachhui, R, Stuehr, D.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2004-06-09 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for isozyme-specific regulation of electron transfer in nitric-oxide synthase
J.Biol.Chem., 279, 2004
|
|
1M9B
 
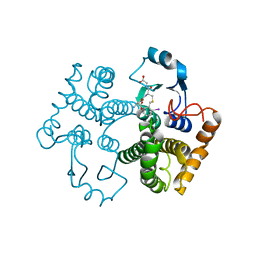 | | Crystal structure of the 26 kDa glutathione S-transferase from Schistosoma japonicum complexed with gamma-glutamyl[S-(2-iodobenzyl)cysteinyl]glycine | | Descriptor: | GAMMA-GLUTAMYL[S-(2-IODOBENZYL)CYSTEINYL]GLYCINE, Glutathione S-Transferase 26 kDa | | Authors: | Cardoso, R.M.F, Daniels, D.S, Bruns, C.M, Tainer, J.A. | | Deposit date: | 2002-07-28 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Characterization of the electrophile
binding site and substrate binding
mode of the 26-kDa glutathione
S-transferase from Schistosoma
japonicum
PROTEINS: STRUCT.,FUNCT.,GENET., 51, 2003
|
|
1M99
 
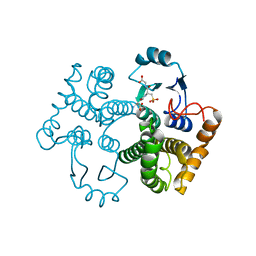 | | Crystal structure of the 26 kDa glutathione S-transferase from Schistosoma japonicum complexed with glutathione sulfonic acid | | Descriptor: | GLUTATHIONE SULFONIC ACID, Glutathione S-Transferase 26kDa | | Authors: | Cardoso, R.M.F, Daniels, D.S, Bruns, C.M, Tainer, J.A. | | Deposit date: | 2002-07-28 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of the electrophile
binding site and substrate binding
mode of the 26-kDa glutathione
S-transferase from Schistosoma
japonicum
PROTEINS: STRUCT.,FUNCT.,GENET., 51, 2003
|
|
1M9A
 
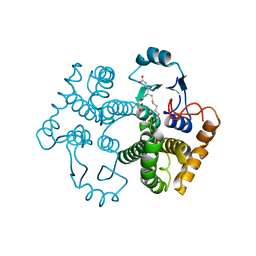 | | Crystal structure of the 26 kDa glutathione S-transferase from Schistosoma japonicum complexed with S-hexylglutathione | | Descriptor: | Glutathione S-Transferase 26 kDa, S-HEXYLGLUTATHIONE | | Authors: | Cardoso, R.M.F, Daniels, D.S, Bruns, C.M, Tainer, J.A. | | Deposit date: | 2002-07-28 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of the electrophile
binding site and substrate binding
mode of the 26-kDa glutathione
S-transferase from Schistosoma
japonicum
PROTEINS: STRUCT.,FUNCT.,GENET., 51, 2003
|
|
