4FE1
 
 | | Improving the Accuracy of Macromolecular Structure Refinement at 7 A Resolution | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Fromme, R, Adams, P.D, Fromme, P, Levitt, M, Schroeder, G.F, Brunger, A.T. | | Deposit date: | 2012-05-29 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.9228 Å) | | Cite: | Improving the accuracy of macromolecular structure refinement at 7 A resolution.
Structure, 20, 2012
|
|
3TX8
 
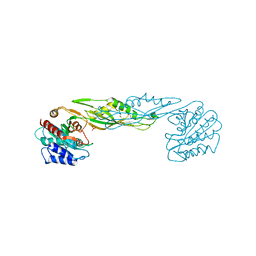 | | Crystal structure of a succinyl-diaminopimelate desuccinylase (ArgE) from Corynebacterium glutamicum ATCC 13032 at 2.97 A resolution | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Succinyl-diaminopimelate desuccinylase | | Authors: | Joint Center for Structural Genomics (JCSG), Brunger, A.T, Terwilliger, T.C, Read, R.J, Adams, P.D, Levitt, M, Schroder, G.F. | | Deposit date: | 2011-09-22 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.972 Å) | | Cite: | Application of DEN refinement and automated model building to a difficult case of molecular-replacement phasing: the structure of a putative succinyl-diaminopimelate desuccinylase from Corynebacterium glutamicum.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1BAF
 
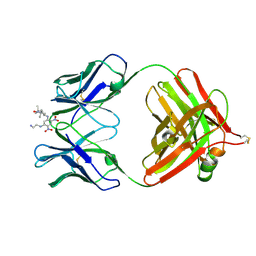 | | 2.9 ANGSTROMS RESOLUTION STRUCTURE OF AN ANTI-DINITROPHENYL-SPIN-LABEL MONOCLONAL ANTIBODY FAB FRAGMENT WITH BOUND HAPTEN | | Descriptor: | IGG1-KAPPA AN02 FAB (HEAVY CHAIN), IGG1-KAPPA AN02 FAB (LIGHT CHAIN), N-(2-AMINO-ETHYL)-4,6-DINITRO-N'-(2,2,6,6-TETRAMETHYL-1-OXY-PIPERIDIN-4-YL)-BENZENE-1,3-DIAMINE | | Authors: | Leahy, D.J, Brunger, A.T, Fox, R.O, Hynes, T.R. | | Deposit date: | 1992-01-16 | | Release date: | 1994-01-31 | | Last modified: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | 2.9 A resolution structure of an anti-dinitrophenyl-spin-label monoclonal antibody Fab fragment with bound hapten.
J.Mol.Biol., 221, 1991
|
|
2ISH
 
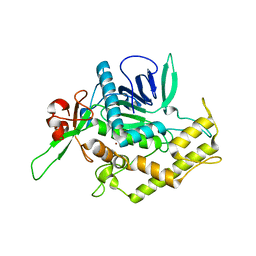 | | Botulinum Neurotoxin A Light Chain WT Crystal Form C | | Descriptor: | Neurotoxin BoNT/A, ZINC ION | | Authors: | Brunger, A.T, Stegmann, C.M. | | Deposit date: | 2006-10-17 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of metalloprotease botulinum serotype A from a pseudo-peptide binding mode to a small molecule that is active in primary neurons.
J.Biol.Chem., 282, 2007
|
|
2ISE
 
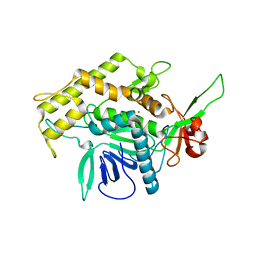 | | Botulinum Neurotoxin A Light Chain WT Crystal Form A | | Descriptor: | Neurotoxin BoNT/A, ZINC ION | | Authors: | Brunger, A.T, Stegmann, C.M. | | Deposit date: | 2006-10-17 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibition of metalloprotease botulinum serotype A from a pseudo-peptide binding mode to a small molecule that is active in primary neurons.
J.Biol.Chem., 282, 2007
|
|
2ISG
 
 | | Botulinum Neurotoxin A Light Chain WT Crystal Form B | | Descriptor: | NICKEL (II) ION, Neurotoxin BoNT/A, ZINC ION | | Authors: | Brunger, A.T, Stegmann, C.M. | | Deposit date: | 2006-10-17 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of metalloprotease botulinum serotype A from a pseudo-peptide binding mode to a small molecule that is active in primary neurons.
J.Biol.Chem., 282, 2007
|
|
8CZI
 
 | |
4WMG
 
 | | Structure of hen egg-white lysozyme from a microfludic harvesting device using synchrotron radiation (2.5A) | | Descriptor: | Lysozyme C | | Authors: | Lyubimov, A.Y, Murray, T.D, Koehl, A, Uervirojnangkoorn, M, Zeldin, O.B, Cohen, A.E, Soltis, S.M, Baxter, E.M, Brewster, A.S, Sauter, N.K, Brunger, A.T, Berger, J.M. | | Deposit date: | 2014-10-08 | | Release date: | 2015-04-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Capture and X-ray diffraction studies of protein microcrystals in a microfluidic trap array.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8FA2
 
 | |
8FA1
 
 | |
4QQP
 
 | | Crystal structure of C1QL3 mutant D207A | | Descriptor: | CADMIUM ION, Complement C1q-like protein 3 | | Authors: | Ressl, S, Brunger, A.T. | | Deposit date: | 2014-06-27 | | Release date: | 2015-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.464 Å) | | Cite: | Structures of C1q-like Proteins Reveal Unique Features among the C1q/TNF Superfamily.
Structure, 23, 2015
|
|
4QQO
 
 | | Crystal structure of C1QL3 mutant D205A | | Descriptor: | Complement C1q-like protein 3, MAGNESIUM ION | | Authors: | Ressl, S, Brunger, A.T. | | Deposit date: | 2014-06-27 | | Release date: | 2015-04-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.031 Å) | | Cite: | Structures of C1q-like Proteins Reveal Unique Features among the C1q/TNF Superfamily.
Structure, 23, 2015
|
|
4QQL
 
 | | Crystal structure of C1QL3 in P1 space group | | Descriptor: | Complement C1q-like protein 3, MAGNESIUM ION | | Authors: | Ressl, S, Brunger, A.T. | | Deposit date: | 2014-06-27 | | Release date: | 2015-04-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structures of C1q-like Proteins Reveal Unique Features among the C1q/TNF Superfamily.
Structure, 23, 2015
|
|
4QQ2
 
 | | Crystal structure of C1QL1 | | Descriptor: | C1q-related factor, CADMIUM ION | | Authors: | Ressl, S, Brunger, A.T. | | Deposit date: | 2014-06-26 | | Release date: | 2015-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structures of C1q-like Proteins Reveal Unique Features among the C1q/TNF Superfamily.
Structure, 23, 2015
|
|
4QPY
 
 | | Crystal structure of C1QL2 | | Descriptor: | Complement C1q-like protein 2 | | Authors: | Ressl, S, Brunger, A.T. | | Deposit date: | 2014-06-25 | | Release date: | 2015-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.384 Å) | | Cite: | Structures of C1q-like Proteins Reveal Unique Features among the C1q/TNF Superfamily.
Structure, 23, 2015
|
|
4QQH
 
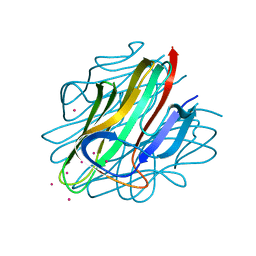 | | Crystal structure of C1QL3 in space group H32 | | Descriptor: | CADMIUM ION, Complement C1q-like protein 3 | | Authors: | Ressl, S, Brunger, A.T. | | Deposit date: | 2014-06-27 | | Release date: | 2015-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structures of C1q-like Proteins Reveal Unique Features among the C1q/TNF Superfamily.
Structure, 23, 2015
|
|
7RZS
 
 | |
7RZT
 
 | |
7RZU
 
 | |
7RZR
 
 | |
7RZV
 
 | |
7RZQ
 
 | |
7TIK
 
 | |
5CCI
 
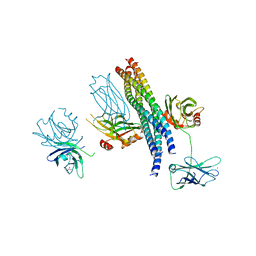 | | Structure of the Mg2+-bound synaptotagmin-1 SNARE complex (short unit cell form) | | Descriptor: | MAGNESIUM ION, Synaptosomal-associated protein 25, Synaptotagmin-1, ... | | Authors: | Zhou, Q, Zhao, M, Lyubimov, A.Y, Uervirojnangkoorn, M, Weis, W.I, Brunger, A.T. | | Deposit date: | 2015-07-02 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Architecture of the synaptotagmin-SNARE machinery for neuronal exocytosis.
Nature, 525, 2015
|
|
5CCH
 
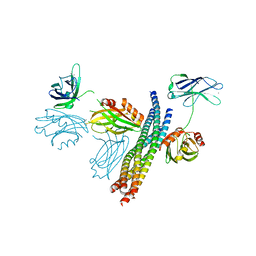 | | Structure of the Ca2+-bound synaptotagmin-1 SNARE complex (short unit cell form) | | Descriptor: | CALCIUM ION, Synaptosomal-associated protein 25, Synaptotagmin-1, ... | | Authors: | Zhou, Q, Zhao, M, Lyubimov, A.Y, Uervirojnangkoorn, M, Zeldin, O.B, Weis, W.I, Brunger, A.T. | | Deposit date: | 2015-07-02 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Architecture of the synaptotagmin-SNARE machinery for neuronal exocytosis.
Nature, 525, 2015
|
|
