8F8E
 
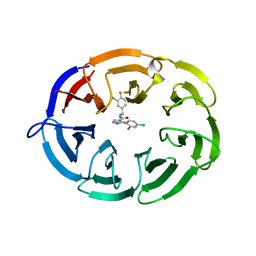 | | Crystal structure of the WDR domain of human DCAF1 in complex with OICR-8268 compound | | Descriptor: | (3P)-N-[(1S)-3-amino-1-(3-chloro-4-fluorophenyl)-3-oxopropyl]-3-(4-chloro-2-fluorophenyl)-1H-pyrazole-4-carboxamide, CITRIC ACID, DDB1- and CUL4-associated factor 1 | | Authors: | Kimani, S, Li, A, Dong, A, Li, Y, Hutchinson, A, Seitova, A, Wilson, B, Al-Awar, R, Vedadi, M, Brown, P, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-21 | | Release date: | 2023-03-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the WDR domain of human DCAF1 in complex with OICR-8268 compound
To be published
|
|
4BUL
 
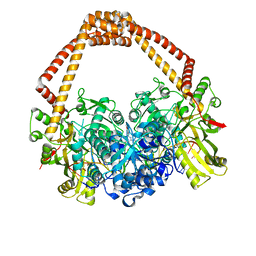 | | Novel hydroxyl tricyclics (e.g. GSK966587) as potent inhibitors of bacterial type IIA topoisomerases | | Descriptor: | (S)-4-((4-(((2,3-dihydro-[1,4]dioxino[2,3-c]pyridin-7-yl)methyl)amino)piperidin-1-yl)methyl)-3-fluoro-4-hydroxy-4H-pyrrolo[3,2,1-de][1,5]naphthyridin-7(5H)-one, 5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*TP*AP*CP*AP*CP *CP*GP*CP*AP*C)-3', 5'-D(*TP*GP*TP*GP*CP*GP*GP*TP*GP*TP*AP*CP*CP*TP *AP*CP*GP*GP*CP*T)-3', ... | | Authors: | Miles, T.J, Hennessy, A.J, Bax, B, Brooks, G, Brown, B.S, Brown, P, Cailleau, N, Chen, D, Dabbs, S, Davies, D.T, Esken, J.M, Giordano, I, Hoover, J.L, Huang, J, Jones, G.E, Sukmar, S.K.K, Spitzfaden, C, Markwell, R.E, Minthorn, E.A, Rittenhouse, S, Gwynn, M.N, Pearson, N.D. | | Deposit date: | 2013-06-20 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel Hydroxyl Tricyclics (E.G., Gsk966587) as Potent Inhibitors of Bacterial Type Iia Topoisomerases.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2A2K
 
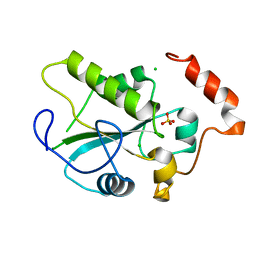 | | Crystal Structure of an active site mutant, C473S, of Cdc25B Phosphatase Catalytic Domain | | Descriptor: | CHLORIDE ION, M-phase inducer phosphatase 2, SULFATE ION | | Authors: | Sohn, J, Parks, J, Buhrman, G, Brown, P, Kristjansdottir, K, Safi, A, Yang, W, Edelsbrunner, H, Rudolph, J. | | Deposit date: | 2005-06-22 | | Release date: | 2006-01-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Experimental Validation of the Docking Orientation of Cdc25 with Its Cdk2-CycA Protein Substrate.
Biochemistry, 44, 2005
|
|
7UFV
 
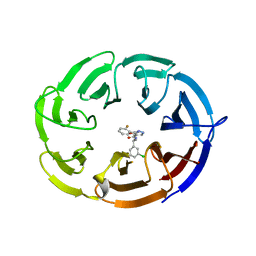 | | Crystal structure of the WDR domain of human DCAF1 in complex with OICR-6766 | | Descriptor: | (3P)-N-[(1S)-3-amino-1-(3-chlorophenyl)-3-oxopropyl]-3-(2-fluorophenyl)-1H-pyrazole-4-carboxamide, DDB1- and CUL4-associated factor 1, UNKNOWN ATOM OR ION | | Authors: | Kimani, S, Li, A, Li, Y, Dong, A, Hutchinson, A, Seitova, A, Wilson, B, Al-Awar, R, Vedadi, M, Brown, P, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-23 | | Release date: | 2022-05-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Nanomolar DCAF1 Small Molecule Ligands.
J.Med.Chem., 66, 2023
|
|
5IWI
 
 | | 1.98A structure of GSK945237 with S.aureus DNA gyrase and singly nicked DNA | | Descriptor: | (1R)-1-[(4-{[(6,7-dihydro[1,4]dioxino[2,3-c]pyridazin-3-yl)methyl]amino}piperidin-1-yl)methyl]-9-fluoro-1,2-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-4-one, CHLORIDE ION, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*TP*AP*CP*AP*CP*CP*GP*CP*AP*CP*A)-3'), ... | | Authors: | Bax, B.D, Miles, T.J. | | Deposit date: | 2016-03-22 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Novel tricyclics (e.g., GSK945237) as potent inhibitors of bacterial type IIA topoisomerases.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5IWM
 
 | | 2.5A structure of GSK945237 with S.aureus DNA gyrase and DNA. | | Descriptor: | (1R)-1-[(4-{[(6,7-dihydro[1,4]dioxino[2,3-c]pyridazin-3-yl)methyl]amino}piperidin-1-yl)methyl]-9-fluoro-1,2-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-4-one, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*TP*TP*CP*AP*CP*CP*GP*CP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*GP*CP*GP*GP*TP*GP*AP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Bax, B.D, Miles, T.J. | | Deposit date: | 2016-03-22 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel tricyclics (e.g., GSK945237) as potent inhibitors of bacterial type IIA topoisomerases.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5HZM
 
 | | Human HMT1 hnRNP methyltransferase-like protein 6 (S. cerevisiae) | | Descriptor: | Protein arginine N-methyltransferase 6, S-ADENOSYL-L-HOMOCYSTEINE, UNKNOWN ATOM OR ION | | Authors: | DONG, A, ZENG, H, WALKER, J.R, Seitova, A, Hutchinson, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, MIN, J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-02-02 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis of arginine asymmetrical dimethylation by PRMT6.
Biochem. J., 473, 2016
|
|
4QQK
 
 | | Human HMT1 hnRNP methyltransferase-like protein 6 (S. cerevisiae) with GMS | | Descriptor: | (5S)-5-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}-N~6~-carbamimidoyl-L-lysine, GLYCEROL, Protein arginine N-methyltransferase 6, ... | | Authors: | Dong, A, Zeng, H, He, H, Wernimont, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Min, J, Luo, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural basis of arginine asymmetrical dimethylation by PRMT6.
Biochem. J., 473, 2016
|
|
1JIL
 
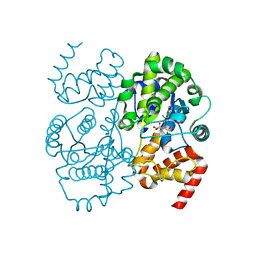 | | Crystal structure of S. aureus TyrRS in complex with SB284485 | | Descriptor: | [2-AMINO-3-(4-HYDROXY-PHENYL)-PROPIONYLAMINO]- (3,4,5-TRIHYDROXY-6-METHYL-TETRAHYDRO-PYRAN-2-YL)- ACETIC ACID, tyrosyl-tRNA synthetase | | Authors: | Qiu, X, Janson, C.A, Smith, W.W, Jarvest, R.L. | | Deposit date: | 2001-07-02 | | Release date: | 2001-10-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Staphylococcus aureus tyrosyl-tRNA synthetase in complex with a class of potent and specific inhibitors.
Protein Sci., 10, 2001
|
|
1JIJ
 
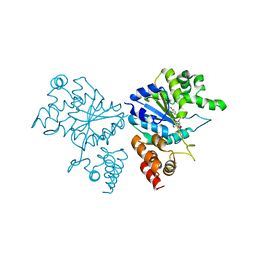 | | Crystal structure of S. aureus TyrRS in complex with SB-239629 | | Descriptor: | [2-AMINO-3-(4-HYDROXY-PHENYL)-PROPIONYLAMINO]-(1,3,4,5-TETRAHYDROXY-4-HYDROXYMETHYL-PIPERIDIN-2-YL)- ACETIC ACID, tyrosyl-tRNA synthetase | | Authors: | Qiu, X, Janson, C.A, Smith, W.W, Jarvest, R.L. | | Deposit date: | 2001-07-02 | | Release date: | 2001-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of Staphylococcus aureus tyrosyl-tRNA synthetase in complex with a class of potent and specific inhibitors.
Protein Sci., 10, 2001
|
|
1JIK
 
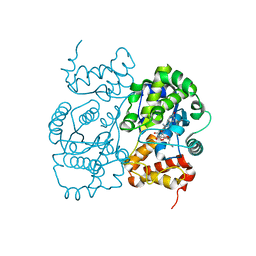 | | Crystal structure of S. aureus TyrRS in complex with SB-243545 | | Descriptor: | [2-AMINO-3-(4-HYDROXY-PHENYL)-PROPIONYLAMINO]-(1,3,4,5-TETRAHYDROXY-4-HYDROXYMETHYL-PIPERIDIN-2-YL)- ACETIC ACID BUTYL ESTER, tyrosyl-tRNA synthetase | | Authors: | Qiu, X, Janson, C.A, Smith, W.W, Jarvest, R.L. | | Deposit date: | 2001-07-02 | | Release date: | 2001-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Staphylococcus aureus tyrosyl-tRNA synthetase in complex with a class of potent and specific inhibitors.
Protein Sci., 10, 2001
|
|
1JII
 
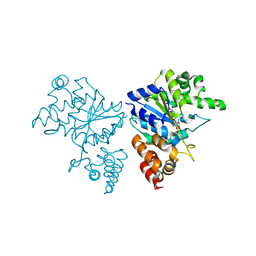 | | Crystal structure of S. aureus TyrRS in complex with SB-219383 | | Descriptor: | [2-AMINO-3-(4-HYDROXY-PHENYL)-PROPIONYLAMINO]- (2,4,5,8-TETRAHYDROXY-7-OXA-2-AZA-BICYCLO[3.2.1]OCT-3-YL)- ACETIC ACID, tyrosyl-tRNA synthetase | | Authors: | Qiu, X, Janson, C.A, Smith, W.W, Jarvest, R.L. | | Deposit date: | 2001-07-02 | | Release date: | 2001-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of Staphylococcus aureus tyrosyl-tRNA synthetase in complex with a class of potent and specific inhibitors.
Protein Sci., 10, 2001
|
|
2MTG
 
 | | Solution structure of the RRM1 of human LARP6 | | Descriptor: | La-related protein 6 | | Authors: | Martino, L, Atkinson, A.R, Kelly, G, Conte, M.R. | | Deposit date: | 2014-08-18 | | Release date: | 2014-12-24 | | Last modified: | 2015-01-28 | | Method: | SOLUTION NMR | | Cite: | Synergic interplay of the La motif, RRM1 and the interdomain linker of LARP6 in the recognition of collagen mRNA expands the RNA binding repertoire of the La module.
Nucleic Acids Res., 43, 2015
|
|
2MTF
 
 | | Solution structure of the La motif of human LARP6 | | Descriptor: | La-related protein 6 | | Authors: | Martino, L, Salisbury, N.JH, Atkinson, A.R, Kelly, G, Conte, M.R. | | Deposit date: | 2014-08-18 | | Release date: | 2014-12-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Synergic interplay of the La motif, RRM1 and the interdomain linker of LARP6 in the recognition of collagen mRNA expands the RNA binding repertoire of the La module.
Nucleic Acids Res., 43, 2015
|
|
1GWX
 
 | | MOLECULAR RECOGNITION OF FATTY ACIDS BY PEROXISOME PROLIFERATOR-ACTIVATED RECEPTORS | | Descriptor: | 2-(4-{3-[1-[2-(2-CHLORO-6-FLUORO-PHENYL)-ETHYL]-3-(2,3-DICHLORO-PHENYL)-UREIDO]-PROPYL}-PHENOXY)-2-METHYL-PROPIONIC ACID, PROTEIN (PPAR-DELTA) | | Authors: | Xu, H.E, Lambert, M.H, Montana, V.G, Park, D.J, Blanchard, S, Brown, P, Sternbach, D, Lehmann, J, Bruce, G.W, Willson, T.M, Kliewer, S.A, Milburn, M.V. | | Deposit date: | 1999-03-17 | | Release date: | 2000-03-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular recognition of fatty acids by peroxisome proliferator-activated receptors.
Mol.Cell, 3, 1999
|
|
4HC4
 
 | | Human HMT1 hnRNP methyltransferase-like protein 6 (S. cerevisiae) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein arginine N-methyltransferase 6, ... | | Authors: | Dong, A, Zeng, H, He, H, El Bakkouri, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis of arginine asymmetrical dimethylation by PRMT6.
Biochem. J., 473, 2016
|
|
3UR4
 
 | | Crystal structure of human WD repeat domain 5 with compound | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Dong, A, Dombrovski, L, Senisterra, G, Wernimont, A, Wasney, G.A, Allali Hassani, A, Nguyen, K.T, Smil, D, Bolshan, Y, Hajian, T, Poda, G, Chau, I, Al-Awar, R, Bountra, C, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Brown, P, Schapira, M, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-11-21 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5.
Biochem. J., 449, 2013
|
|
2GWX
 
 | | MOLECULAR RECOGNITION OF FATTY ACIDS BY PEROXISOME PROLIFERATOR-ACTIVATED RECEPTORS | | Descriptor: | PROTEIN (PPAR-DELTA) | | Authors: | Xu, H.E, Lambert, M.H, Montana, V.G, Park, D.J, Blanchard, S, Brown, P, Sternbach, D, Lehmann, J, Bruce, G.W, Willson, T.M, Kliewer, S.A, Milburn, M.V. | | Deposit date: | 1999-03-11 | | Release date: | 2000-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular recognition of fatty acids by peroxisome proliferator-activated receptors.
Mol.Cell, 3, 1999
|
|
4IO5
 
 | |
4IO4
 
 | | Crystal Structure of the AvGluR1 ligand binding domain complex with serine at 1.94 Angstrom resolution | | Descriptor: | AvGluR1 ligand binding domain, CHLORIDE ION, GLYCEROL, ... | | Authors: | Lomash, S, Chittori, S, Mayer, M.L. | | Deposit date: | 2013-01-07 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.941 Å) | | Cite: | Anions Mediate Ligand Binding in Adineta vaga Glutamate Receptor Ion Channels.
Structure, 21, 2013
|
|
4IO6
 
 | |
4IO7
 
 | |
4IO3
 
 | |
4IO2
 
 | |
2UZQ
 
 | | Protein Phosphatase, New Crystal Form | | Descriptor: | M-PHASE INDUCER PHOSPHATASE 2, PHOSPHATE ION | | Authors: | Hillig, R.C, Eberspaecher, U. | | Deposit date: | 2007-05-01 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | New Crystal Form of Protein Phosphatase Cdc25B Triggered by Guanidinium Chloride as an Additive
To be Published
|
|
