1ELS
 
 | | CATALYTIC METAL ION BINDING IN ENOLASE: THE CRYSTAL STRUCTURE OF ENOLASE-MN2+-PHOSPHONOACETOHYDROXAMATE COMPLEX AT 2.4 ANGSTROMS RESOLUTION | | Descriptor: | ENOLASE, MANGANESE (II) ION, PHOSPHONOACETOHYDROXAMIC ACID | | Authors: | Zhang, E, Hatada, M, Brewer, J.M, Lebioda, L. | | Deposit date: | 1994-04-05 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Catalytic metal ion binding in enolase: the crystal structure of an enolase-Mn2+-phosphonoacetohydroxamate complex at 2.4-A resolution.
Biochemistry, 33, 1994
|
|
2AKZ
 
 | | Fluoride Inhibition of Enolase: Crystal Structure of the Inhibitory Complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FLUORIDE ION, Gamma enolase, ... | | Authors: | Qin, J, Chai, G, Brewer, J.M, Lovelace, L.L. | | Deposit date: | 2005-08-04 | | Release date: | 2006-03-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Fluoride inhibition of enolase: crystal structure and thermodynamics
Biochemistry, 45, 2006
|
|
2AKM
 
 | | Fluoride Inhibition of Enolase: Crystal Structure of the Inhibitory Complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Gamma enolase, MAGNESIUM ION, ... | | Authors: | Qin, J, Chai, G, Brewer, J.M, Lovelace, L.L. | | Deposit date: | 2005-08-03 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Fluoride inhibition of enolase: crystal structure and thermodynamics
Biochemistry, 45, 2006
|
|
1FPM
 
 | | MONOVALENT CATION BINDING SITES IN N10-FORMYLTETRAHYDROFOLATE SYNTHETASE FROM MOORELLA THERMOACETICA | | Descriptor: | CESIUM ION, FORMATE--TETRAHYDROFOLATE LIGASE, SULFATE ION | | Authors: | Radfar, R, Leaphart, A, Brewer, J.M, Minor, W, Odom, J.D. | | Deposit date: | 2000-08-31 | | Release date: | 2001-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cation binding and thermostability of FTHFS monovalent cation binding sites and thermostability of N10-formyltetrahydrofolate synthetase from Moorella thermoacetica.
Biochemistry, 39, 2000
|
|
1FP7
 
 | | MONOVALENT CATION BINDING SITES IN N10-FORMYLTETRAHYDROFOLATE SYNTHETASE FROM MOORELLA THERMOACETICA | | Descriptor: | FORMATE--TETRAHYDROFOLATE LIGASE, POTASSIUM ION, SULFATE ION | | Authors: | Radfar, R, Leaphart, A, Brewer, J.M, Minor, W, Odom, J.D. | | Deposit date: | 2000-08-30 | | Release date: | 2001-08-30 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Cation binding and thermostability of FTHFS monovalent cation binding sites and thermostability of N10-formyltetrahydrofolate synthetase from Moorella thermoacetica.
Biochemistry, 39, 2000
|
|
7ENL
 
 | |
1NEL
 
 | | FLUORIDE INHIBITION OF YEAST ENOLASE: CRYSTAL STRUCTURE OF THE ENOLASE-MG2+-F--PI COMPLEX AT 2.6-ANGSTROMS RESOLUTION | | Descriptor: | ENOLASE, FLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Lebioda, L, Zhang, E, Lewinski, K, Brewer, M.J. | | Deposit date: | 1993-08-20 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Fluoride inhibition of yeast enolase: crystal structure of the enolase-Mg(2+)-F(-)-Pi complex at 2.6 A resolution.
Proteins, 16, 1993
|
|
4ENL
 
 | |
5ENL
 
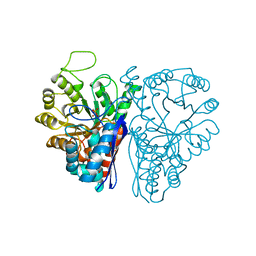 | |
6ENL
 
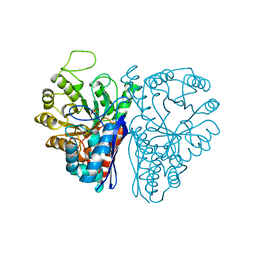 | |
3UCD
 
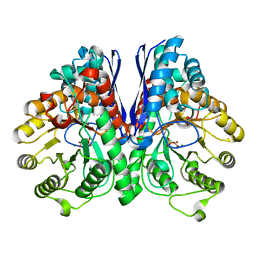 | | Asymmetric complex of human neuron specific enolase-2-PGA/PEP | | Descriptor: | 2-PHOSPHOGLYCERIC ACID, Gamma-enolase, MAGNESIUM ION, ... | | Authors: | Qin, J, Chai, G, Brewer, J, Lovelace, L, Lebioda, L. | | Deposit date: | 2011-10-26 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structures of asymmetric complexes of human neuron specific enolase with resolved substrate and product and an analogous complex with two inhibitors indicate subunit interaction and inhibitor cooperativity.
J.Inorg.Biochem., 111, 2012
|
|
3UJS
 
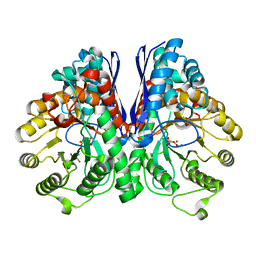 | | Asymmetric complex of human neuron specific enolase-6-PGA/PEP | | Descriptor: | (2R)-2-(phosphonooxy)propanoic acid, (2R)-3-oxo-2-(phosphonooxy)propanoic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Qin, J, Chai, G, Brewer, J, Lovelace, L, Lebioda, L. | | Deposit date: | 2011-11-08 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of asymmetric complexes of human neuron specific enolase with resolved substrate and product and an analogous complex with two inhibitors indicate subunit interaction and inhibitor cooperativity.
J.Inorg.Biochem., 111, 2012
|
|
3UJE
 
 | | Asymmetric complex of human neuron specific enolase-3-PGA/PEP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-PHOSPHOGLYCERIC ACID, Gamma-enolase, ... | | Authors: | Qin, J, Chai, G, Brewer, J, Lovelace, L, Lebioda, L. | | Deposit date: | 2011-11-07 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of asymmetric complexes of human neuron specific enolase with resolved substrate and product and an analogous complex with two inhibitors indicate subunit interaction and inhibitor cooperativity.
J.Inorg.Biochem., 111, 2012
|
|
3UCC
 
 | | Asymmetric complex of human neuron specific enolase-1-PGA/PEP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-PHOSPHOGLYCERIC ACID, Gamma-enolase, ... | | Authors: | Qin, J, Chai, G, Brewer, J, Lovelace, L, Lebioda, L. | | Deposit date: | 2011-10-26 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of asymmetric complexes of human neuron specific enolase with resolved substrate and product and an analogous complex with two inhibitors indicate subunit interaction and inhibitor cooperativity.
J.Inorg.Biochem., 111, 2012
|
|
3UJF
 
 | | Asymmetric complex of human neuron specific enolase-4-PGA/PEP | | Descriptor: | 2-PHOSPHOGLYCERIC ACID, Gamma-enolase, MAGNESIUM ION, ... | | Authors: | Qin, J, Chai, G, Brewer, J, Lovelace, L, Lebioda, L. | | Deposit date: | 2011-11-07 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of asymmetric complexes of human neuron specific enolase with resolved substrate and product and an analogous complex with two inhibitors indicate subunit interaction and inhibitor cooperativity.
J.Inorg.Biochem., 111, 2012
|
|
3UJR
 
 | | Asymmetric complex of human neuron specific enolase-5-PGA/PEP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-PHOSPHOGLYCERIC ACID, Gamma-enolase, ... | | Authors: | Qin, J, Chai, G, Brewer, J, Lovelace, L, Lebioda, L. | | Deposit date: | 2011-11-08 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of asymmetric complexes of human neuron specific enolase with resolved substrate and product and an analogous complex with two inhibitors indicate subunit interaction and inhibitor cooperativity.
J.Inorg.Biochem., 111, 2012
|
|
2ONE
 
 | |
3ENL
 
 | |
