5ONM
 
 | | Crystal Structure of Ectoine Synthase from P. lautus | | Descriptor: | FE (III) ION, L-ectoine synthase, SULFATE ION | | Authors: | Bremer, E. | | Deposit date: | 2017-08-04 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Illuminating the catalytic core of ectoine synthase through structural and biochemical analysis.
Sci Rep, 9, 2019
|
|
5ONN
 
 | | Crystal Structure of Ectoine Synthase from P. lautus | | Descriptor: | (2~{S})-4-acetamido-2-azanyl-butanoic acid, FE (III) ION, L-ectoine synthase | | Authors: | Bremer, E. | | Deposit date: | 2017-08-04 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Illuminating the catalytic core of ectoine synthase through structural and biochemical analysis.
Sci Rep, 9, 2019
|
|
5ONO
 
 | | Crystal Structure of Ectoine Synthase from P. lautus | | Descriptor: | (4S)-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, FE (III) ION, L-ectoine synthase | | Authors: | Bremer, E. | | Deposit date: | 2017-08-04 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Illuminating the catalytic core of ectoine synthase through structural and biochemical analysis.
Sci Rep, 9, 2019
|
|
3HCQ
 
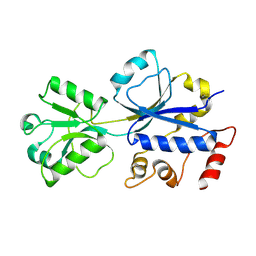 | | Structural analysis of the choline binding protein ChoX in a semi-closed and ligand-free conformation | | Descriptor: | Putative choline ABC transporter, periplasmic solute-binding component | | Authors: | Oswald, C, Smits, S.H.J, Hoeing, M, Bremer, E, Schmitt, L. | | Deposit date: | 2009-05-06 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural analysis of the choline-binding protein ChoX in a semi-closed and ligand-free conformation.
Biol.Chem., 390, 2009
|
|
3MAM
 
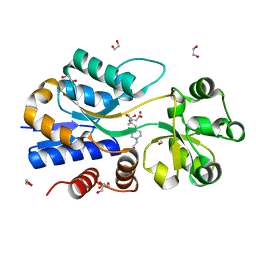 | | A molecular switch changes the low to the high affinity state in the substrate binding protein AfProX | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Osmoprotection protein (ProX), ... | | Authors: | Tschapek, B, Pittelkow, M, Bremer, E, Schmitt, L, Smits, S.H. | | Deposit date: | 2010-03-24 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Arg149 Is Involved in Switching the Low Affinity, Open State of the Binding Protein AfProX into Its High Affinity, Closed State.
J.Mol.Biol., 411, 2011
|
|
5NXY
 
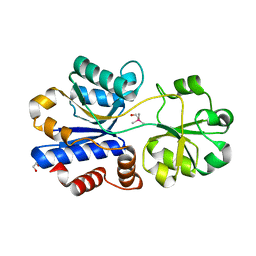 | | Crystal structure of OpuAC from B. subtilis in complex with Arsenobetaine | | Descriptor: | 1,2-ETHANEDIOL, 2-(trimethyl-lambda~5~-arsanyl)ethanol, Osmotically activated L-carnitine/choline ABC transporter substrate-binding protein OpuCC | | Authors: | Hofmann, T, Bremer, E, Schmitt, L, Smits, S. | | Deposit date: | 2017-05-11 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Arsenobetaine: an ecophysiologically important organoarsenical confers cytoprotection against osmotic stress and growth temperature extremes.
Environ. Microbiol., 20, 2018
|
|
5NXX
 
 | | Crystal structure of OpuAC from B. subtilis in complex with Arsenobetaine | | Descriptor: | (trimethylarsonio)acetate, Glycine betaine ABC transport system glycine betaine-binding protein OpuAC | | Authors: | Hofmann, T, Bremer, E, Schmitt, L, Smits, S. | | Deposit date: | 2017-05-11 | | Release date: | 2018-03-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Arsenobetaine: an ecophysiologically important organoarsenical confers cytoprotection against osmotic stress and growth temperature extremes.
Environ. Microbiol., 20, 2018
|
|
3R6U
 
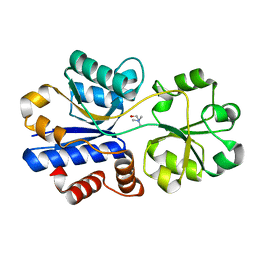 | | Crystal structure of choline binding protein OpuBC from Bacillus subtilis | | Descriptor: | CHOLINE ION, Choline-binding protein | | Authors: | Pittelkow, M, Tschapek, B, Smits, S.H.J, Schmitt, L, Bremer, E. | | Deposit date: | 2011-03-22 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | The Crystal Structure of the Substrate-Binding Protein OpuBC from Bacillus subtilis in Complex with Choline.
J.Mol.Biol., 411, 2011
|
|
5BXX
 
 | | Crystal structure of the ectoine synthase from the cold-adapted marine bacterium Sphingopyxis alaskensis | | Descriptor: | L-ectoine synthase | | Authors: | Widderich, N, Kobus, S, Hoeppner, A, Bremer, E, Smits, S.H.J. | | Deposit date: | 2015-06-09 | | Release date: | 2016-04-27 | | Last modified: | 2016-07-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemistry and Crystal Structure of Ectoine Synthase: A Metal-Containing Member of the Cupin Superfamily.
Plos One, 11, 2016
|
|
5BY5
 
 | | High resolution structure of the ectoine synthase from the cold-adapted marine bacterium Sphingopyxis alaskensis | | Descriptor: | L-ectoine synthase, S-1,2-PROPANEDIOL | | Authors: | Widderich, N, Kobus, S, Hoeppner, A, Bremer, E, Smits, S.H.J. | | Deposit date: | 2015-06-10 | | Release date: | 2016-04-27 | | Last modified: | 2016-07-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Biochemistry and Crystal Structure of Ectoine Synthase: A Metal-Containing Member of the Cupin Superfamily.
Plos One, 11, 2016
|
|
2B4L
 
 | | Crystal structure of the binding protein OpuAC in complex with glycine betaine | | Descriptor: | 1,2-ETHANEDIOL, Glycine betaine-binding protein, TRIMETHYL GLYCINE | | Authors: | Horn, C, Sohn-Boesser, L, Breed, J, Welte, W, Schmitt, L, Bremer, E. | | Deposit date: | 2005-09-26 | | Release date: | 2006-03-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Determinants for Substrate Specificity of the Ligand-binding Protein OpuAC from Bacillus subtilis for the Compatible Solutes Glycine Betaine and Proline Betaine.
J.Mol.Biol., 357, 2006
|
|
2B4M
 
 | | Crystal structure of the binding protein OpuAC in complex with proline betaine | | Descriptor: | 1,1-DIMETHYL-PROLINIUM, Glycine betaine-binding protein | | Authors: | Horn, C, Sohn-Boesser, L, Breed, J, Welte, W, Schmitt, L, Bremer, E. | | Deposit date: | 2005-09-26 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular Determinants for Substrate Specificity of the Ligand-binding Protein OpuAC from Bacillus subtilis for the Compatible Solutes Glycine Betaine and Proline Betaine.
J.Mol.Biol., 357, 2006
|
|
6EYL
 
 | | Crystal structure of OpuBC in complex with carnitine | | Descriptor: | CARNITINE, Osmotically activated L-carnitine/choline ABC transporter substrate-binding protein OpuCC | | Authors: | Peherstorfer, S, Teichmann, L, Smits, S.H, Sschmitt, L, Bremer, E. | | Deposit date: | 2017-11-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Reprogramming the substrate specificity of an ABC import system by a single amino acid substitution in its cognate ligand binding protein
To Be Published
|
|
6EYG
 
 | | Structure of a OpuBC mutant with bound Glycine betaine | | Descriptor: | Osmotically activated L-carnitine/choline ABC transporter substrate-binding protein OpuCC, TRIMETHYL GLYCINE | | Authors: | Peherstorfer, S, Teichmann, L, Smits, S.H, Schmitt, L, Bremer, E. | | Deposit date: | 2017-11-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure of a OpuBC mutant with bound Glycine betaine
To Be Published
|
|
6EYQ
 
 | | Crystal structure of a mutated OpuBC in complex with choline | | Descriptor: | CHOLINE ION, Choline-binding protein | | Authors: | Peherstorfer, S, Teichmann, L, Smits, S.H, Schmitt, L, Bremer, E. | | Deposit date: | 2017-11-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a mutated OpuBC in complex with choline
To Be Published
|
|
6EYH
 
 | | Structure of a OpuBC mutant with bound Glycine betaine | | Descriptor: | 3-(dimethyl-lambda~4~-sulfanyl)propanoic acid, Choline binding protein OpuBC | | Authors: | Peherstorfer, S, Teichmann, L, Smits, S.H, Schmitt, L, Bremer, E. | | Deposit date: | 2017-11-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of a OpuBC mutant with bound DMSP
To Be Published
|
|
4MHU
 
 | | Crystal structure of EctD from S. alaskensis with bound Fe | | Descriptor: | Ectoine hydroxylase, FE (III) ION, N-DODECYL-N,N-DIMETHYLGLYCINATE | | Authors: | Widderich, N, Hoeppner, A, Pittelkow, M, Heider, J, Smits, S.H, Bremer, E. | | Deposit date: | 2013-08-30 | | Release date: | 2014-09-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Crystal structure of the ectoine hydroxylase, a snapshot of the active site.
J.Biol.Chem., 289, 2014
|
|
4MHR
 
 | | Crystal structure of EctD from S. alaskensis in its apoform | | Descriptor: | Ectoine hydroxylase | | Authors: | Widderich, N, Hoeppner, A, Pittelkow, M, Heider, J, Smits, S.H, Bremer, E. | | Deposit date: | 2013-08-30 | | Release date: | 2014-09-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the ectoine hydroxylase, a snapshot of the active site.
J.Biol.Chem., 289, 2014
|
|
4NMI
 
 | |
4Q5O
 
 | | Crystal structure of EctD from S. alaskensis with 2-oxoglutarate and 5-hydroxyectoine | | Descriptor: | (4S,5S)-5-HYDROXY-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, 2-OXOGLUTARIC ACID, Ectoine hydroxylase, ... | | Authors: | Hoeppner, A, Widderich, N, Bremer, E, Smits, S.H. | | Deposit date: | 2014-04-17 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of the ectoine hydroxylase, a snapshot of the active site.
J.Biol.Chem., 289, 2014
|
|
3CHG
 
 | | The compatible solute-binding protein OpuAC from Bacillus subtilis in complex with DMSA | | Descriptor: | (dimethyl-lambda~4~-sulfanyl)acetic acid, Glycine betaine-binding protein | | Authors: | Smits, S.H.J, Hoing, M, Lecher, J, Jebbar, M, Schmitt, L, Bremer, E. | | Deposit date: | 2008-03-09 | | Release date: | 2008-08-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Compatible-Solute-Binding Protein OpuAC from Bacillus subtilis: Ligand Binding, Site-Directed Mutagenesis, and Crystallographic Studies
J.Bacteriol., 190, 2008
|
|
3FXB
 
 | | Crystal structure of the ectoine-binding protein UehA | | Descriptor: | (4S)-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, TRAP dicarboxylate transporter, DctP subunit | | Authors: | Lecher, J, Pittelkow, M, Bursy, J, Smits, S.H.J, Schmitt, L, Bremer, E. | | Deposit date: | 2009-01-20 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of UehA in complex with ectoine-A comparison with other TRAP-T binding proteins.
J.Mol.Biol., 389, 2009
|
|
1R9Q
 
 | | structure analysis of ProX in complex with proline betaine | | Descriptor: | 1,1-DIMETHYL-PROLINIUM, Glycine betaine-binding periplasmic protein, UNKNOWN ATOM OR ION | | Authors: | Schiefner, A, Breed, J, Bosser, L, Kneip, S, Gade, J, Holtmann, G, Diederichs, K, Welte, W, Bremer, E. | | Deposit date: | 2003-10-30 | | Release date: | 2004-02-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Cation-pi Interactions as Determinants for Binding of the Compatible Solutes Glycine Betaine and Proline Betaine by the Periplasmic Ligand-binding Protein ProX from Escherichia coli
J.BIOL.CHEM., 279, 2004
|
|
1R9L
 
 | | structure analysis of ProX in complex with glycine betaine | | Descriptor: | Glycine betaine-binding periplasmic protein, TRIMETHYL GLYCINE, UNKNOWN ATOM OR ION | | Authors: | Schiefner, A, Breed, J, Bosser, L, Kneip, S, Gade, J, Holtmann, G, Diederichs, K, Welte, W, Bremer, E. | | Deposit date: | 2003-10-30 | | Release date: | 2004-02-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Cation-pi Interactions as Determinants for Binding of the Compatible Solutes Glycine Betaine and Proline Betaine by the Periplasmic Ligand-binding Protein ProX from Escherichia coli
J.BIOL.CHEM., 279, 2004
|
|
1SW2
 
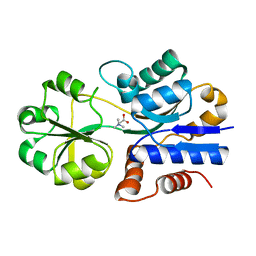 | | Crystal structure of ProX from Archeoglobus fulgidus in complex with glycine betaine | | Descriptor: | TRIMETHYL GLYCINE, osmoprotection protein (proX) | | Authors: | Schiefner, A, Holtmann, G, Diederichs, K, Welte, W, Bremer, E. | | Deposit date: | 2004-03-30 | | Release date: | 2004-09-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the binding of compatible solutes by ProX from the hyperthermophilic archaeon Archaeoglobus fulgidus.
J.Biol.Chem., 279, 2004
|
|
