1PIC
 
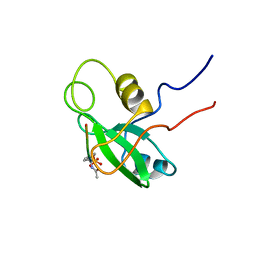 | | PHOSPHATIDYLINOSITOL 3-KINASE, P85-ALPHA SUBUNIT: C-TERMINAL SH2 DOMAIN COMPLEXED WITH A TYR751 PHOSPHOPEPTIDE FROM THE PDGF RECEPTOR, NMR, MINIMIZED MEAN STRUCTURE | | Descriptor: | BETA-PLATELET-DERIVED GROWTH FACTOR RECEPTOR, PHOSPHATIDYLINOSITOL 3-KINASE | | Authors: | Breeze, A.L, Kara, B.V, Barratt, D.G, Anderson, M, Smith, J.C, Luke, R.W, Best, J.R, Cartlidge, S.A. | | Deposit date: | 1997-06-23 | | Release date: | 1997-09-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure of a specific peptide complex of the carboxy-terminal SH2 domain from the p85 alpha subunit of phosphatidylinositol 3-kinase.
EMBO J., 15, 1996
|
|
1H9O
 
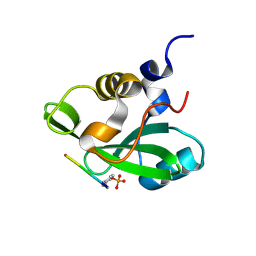 | | PHOSPHATIDYLINOSITOL 3-KINASE, P85-ALPHA SUBUNIT: C-TERMINAL SH2 DOMAIN COMPLEXED WITH A TYR751 PHOSPHOPEPTIDE FROM THE PDGF RECEPTOR, CRYSTAL STRUCTURE AT 1.79 A | | Descriptor: | BETA-PLATELET-DERIVED GROWTH FACTOR RECEPTOR, PHOSPHATIDYLINOSITOL 3-KINASE | | Authors: | Pauptit, R.A, Rowsell, S, Breeze, A.L, Murshudov, G.N, Dennis, C.A, Derbyshire, D.J, Weston, S.A. | | Deposit date: | 2001-03-14 | | Release date: | 2001-03-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | NMR Trial Models: Experiences with the Colicin Immunity Protein Im7 and the P85Alpha C-Terminal Sh2-Peptide Complex
Acta Crystallogr.,Sect.D, 57, 2001
|
|
2PNB
 
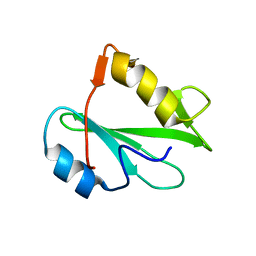 | | STRUCTURE OF AN SH2 DOMAIN OF THE P85 ALPHA SUBUNIT OF PHOSPHATIDYLINOSITOL-3-OH KINASE | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE P85-ALPHA SUBUNIT N-TERMINAL SH2 DOMAIN | | Authors: | Booker, G.W, Breeze, A.L, Downing, A.K, Panayotou, G, Gout, I, Waterfield, M.D, Campbell, I.D. | | Deposit date: | 1992-06-30 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Structure of an SH2 domain of the p85 alpha subunit of phosphatidylinositol-3-OH kinase.
Nature, 358, 1992
|
|
2PNA
 
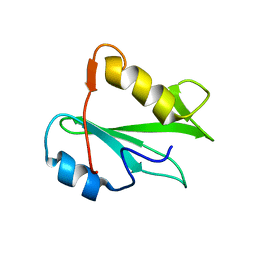 | | STRUCTURE OF AN SH2 DOMAIN OF THE P85 ALPHA SUBUNIT OF PHOSPHATIDYLINOSITOL-3-OH KINASE | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE P85-ALPHA SUBUNIT N-TERMINAL SH2 DOMAIN | | Authors: | Booker, G.W, Breeze, A.L, Downing, A.K, Panayotou, G, Gout, I, Waterfield, M.D, Campbell, I.D. | | Deposit date: | 1992-06-30 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Structure of an SH2 domain of the p85 alpha subunit of phosphatidylinositol-3-OH kinase.
Nature, 358, 1992
|
|
1IMQ
 
 | | COLICIN E9 IMMUNITY PROTEIN IM9, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | IM9 | | Authors: | Osborne, M.J, Breeze, A.L, Lian, L.Y, Reilly, A, James, R, Kleanthous, C, Moore, G.R. | | Deposit date: | 1996-05-30 | | Release date: | 1997-07-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure and 13C nuclear magnetic resonance assignments of the colicin E9 immunity protein Im9.
Biochemistry, 35, 1996
|
|
1IMP
 
 | | COLICIN E9 IMMUNITY PROTEIN IM9, NMR, 21 STRUCTURES | | Descriptor: | IM9 | | Authors: | Osborne, M.J, Breeze, A.L, Lian, L.-Y, Reilly, A, James, R, Kleanthous, C, Moore, G.R. | | Deposit date: | 1996-05-30 | | Release date: | 1997-09-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure and 13C nuclear magnetic resonance assignments of the colicin E9 immunity protein Im9.
Biochemistry, 35, 1996
|
|
4URU
 
 | | The crystal structure of H-Ras and SOS in complex with ligands | | Descriptor: | 4-METHOXY-N-(1,3-THIAZOL-2-YL)BENZENESULFONAMIDE, GTPASE HRAS, SON OF SEVENLESS HOMOLOG 1 | | Authors: | Winter, J.J.G, Anderson, M, Blades, K, Brassington, C, Breeze, A.L, Chresta, C, Embrey, K, Fairley, G, Faulder, P, Finlay, M.R.V, Kettle, J.G, Nowak, T, Overman, R, Patel, S.J, Perkins, P, Spadola, L, Tart, J, Tucker, J, Wrigley, G. | | Deposit date: | 2014-07-02 | | Release date: | 2015-03-04 | | Last modified: | 2015-03-25 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Small Molecule Binding Sites on the Ras:SOS Complex Can be Exploited for Inhibition of Ras Activation.
J.Med.Chem., 58, 2015
|
|
5A4C
 
 | | FGFR1 ligand complex | | Descriptor: | 1,2-ETHANEDIOL, 1-tert-butyl-3-[2-[3-(diethylamino)propylamino]-6-(3,5-dimethoxyphenyl)pyrido[2,3-d]pyrimidin-7-yl]urea, FIBROBLAST GROWTH FACTOR RECEPTOR 1 (FMS-RELATED TYROSINE KINASE 2, ... | | Authors: | Klein, T, Vajpai, N, Phillips, J.J, Davies, G, Holdgate, G.A, Phillips, C, Tucker, J.A, Norman, R.A, Scott, A.S, Higazi, D.R, Lowe, D, Thompson, G.S, Breeze, A.L. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and Dynamic Insights Into the Energetics of Activation Loop Rearrangement in Fgfr1 Kinase.
Nat.Commun., 6, 2015
|
|
5A46
 
 | | FGFR1 in complex with dovitinib | | Descriptor: | 1,2-ETHANEDIOL, 4-amino-5-fluoro-3-[5-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]quinolin-2(1H)-one, FIBROBLAST GROWTH FACTOR RECEPTOR 1 (FMS-RELATED TYROSINE KINASE 2, ... | | Authors: | Klein, T, Vajpai, N, Phillips, J.J, Davies, G, Holdgate, G.A, Phillips, C, Tucker, J.A, Norman, R.A, Scott, A.S, Higazi, D.R, Lowe, D, Thompson, G.S, Breeze, A.L. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural and Dynamic Insights Into the Energetics of Activation Loop Rearrangement in Fgfr1 Kinase.
Nat.Commun., 6, 2015
|
|
5A0O
 
 | | adhiron raised against p300 | | Descriptor: | ADHIRON | | Authors: | Kyle, H.F, Wickson, K.F, Stott, J, Burslem, G.M, Breeze, A.L, Tiede, C, Tomlinson, D.C, Warriner, S.L, Nelson, A, Wilson, A.J, Edwards, T.A. | | Deposit date: | 2015-04-21 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Exploration of the Hif-1Alpha.P300 Interface Using Peptide and Adhiron Phage Display Technologies
Mol.Biosyst., 11, 2015
|
|
2BGE
 
 | | Structure-based design of Protein Tyrosine Phosphatase-1B Inhibitors | | Descriptor: | 1,2,5-THIADIAZOLIDIN-3-ONE-1,1-DIOXIDE, PROTEIN-TYROSINE PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Black, E, Breed, J, Breeze, A.L, Embrey, K, Garcia, R, Gero, T.W, Godfrey, L, Kenny, P.W, Morley, A.D, Minshull, C.A, Pannifer, A.D, Read, J, Rees, A, Russell, D.J, Toader, D, Tucker, J. | | Deposit date: | 2004-12-21 | | Release date: | 2005-05-04 | | Last modified: | 2015-10-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Design of Protein Tyrosine Phosphatase-1B Inhibitors
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2BGD
 
 | | Structure-based design of Protein Tyrosine Phosphatase-1B Inhibitors | | Descriptor: | 5-(4-METHOXYBIPHENYL-3-YL)-1,2,5-THIADIAZOLIDIN-3-ONE 1,1-DIOXIDE, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Black, E, Breed, J, Breeze, A.L, Embrey, K, Garcia, R, Gero, T.W, Godfrey, L, Kenny, P.W, Morley, A.D, Minshull, C.A, Pannifer, A.D, Read, J, Rees, A, Russell, D.J, Toader, D, Tucker, J. | | Deposit date: | 2004-12-21 | | Release date: | 2005-05-04 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Design of Protein Tyrosine Phosphatase-1B Inhibitors
Bioorg.Med.Chem.Lett., 15, 2005
|
|
6YR8
 
 | | Affimer K6 - KRAS protein complex | | Descriptor: | Affimer K6, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Trinh, C.H, Haza, K.Z, Rao, A, Martin, H.L, Tiede, C, Edwards, T.A, McPherson, M.J, Tomlinson, D.C. | | Deposit date: | 2020-04-19 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | RAS-inhibiting biologics identify and probe druggable pockets including an SII-alpha 3 allosteric site.
Nat Commun, 12, 2021
|
|
6YXW
 
 | | Affimer K3 - KRAS protein complex | | Descriptor: | Affimer K3, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Trinh, C.H, Haza, K.Z, Rao, A, Martin, H.L, Tiede, C, Edwards, T.A, McPherson, M.J, Tomlinson, D.C. | | Deposit date: | 2020-05-04 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | RAS-inhibiting biologics identify and probe druggable pockets including an SII-alpha 3 allosteric site.
Nat Commun, 12, 2021
|
|
7NY8
 
 | | Affimer K69 - KRAS protein complex | | Descriptor: | Affimer K69, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Turner, A.L, Trinh, C.H, Haza, K.Z, Rao, A, Martin, H.L, Tiede, C, Edwards, T.A, McPherson, M.J, Tomlinson, D.C. | | Deposit date: | 2021-03-21 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | RAS-inhibiting biologics identify and probe druggable pockets including an SII-alpha 3 allosteric site.
Nat Commun, 12, 2021
|
|
1AJ6
 
 | |
4V04
 
 | | FGFR1 in complex with ponatinib. | | Descriptor: | 1,2-ETHANEDIOL, 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzamide, FIBROBLAST GROWTH FACTOR RECEPTOR 1 (FMS-RELATED TYROSINE KINASE 2, ... | | Authors: | Tucker, J, Klein, T, Breed, J, Breeze, A, Overman, R, Phillips, C, Norman, R.A. | | Deposit date: | 2014-09-10 | | Release date: | 2014-12-10 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural Insights Into Fgfr Kinase Isoform Selectivity: Diverse Binding Modes of Azd4547 and Ponatinib in Complex with Fgfr1 and Fgfr4
Structure, 22, 2014
|
|
4V01
 
 | | FGFR1 in complex with ponatinib (co-crystallisation). | | Descriptor: | 1,2-ETHANEDIOL, 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzamide, FIBROBLAST GROWTH FACTOR RECEPTOR 1 (FMS-RELATED TYROSINE KINASE 2, ... | | Authors: | Tucker, J, Klein, T, Breed, J, Breeze, A, Overman, R, Phillips, C, Norman, R.A. | | Deposit date: | 2014-09-10 | | Release date: | 2014-12-10 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural Insights Into Fgfr Kinase Isoform Selectivity: Diverse Binding Modes of Azd4547 and Ponatinib in Complex with Fgfr1 and Fgfr4
Structure, 22, 2014
|
|
4V05
 
 | | FGFR1 in complex with AZD4547. | | Descriptor: | 1,2-ETHANEDIOL, FIBROBLAST GROWTH FACTOR RECEPTOR 1 (FMS-RELATED TYROSINE KINASE 2, PFEIFFER SYNDROME), ... | | Authors: | Tucker, J, Klein, T, Breed, J, Breeze, A, Overman, R, Phillips, C, Norman, R.A. | | Deposit date: | 2014-09-10 | | Release date: | 2014-12-10 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural Insights Into Fgfr Kinase Isoform Selectivity: Diverse Binding Modes of Azd4547 and Ponatinib in Complex with Fgfr1 and Fgfr4
Structure, 22, 2014
|
|
4UXQ
 
 | | FGFR4 in complex with Ponatinib | | Descriptor: | 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzamide, FIBROBLAST GROWTH FACTOR RECEPTOR 4, SULFATE ION | | Authors: | Tucker, J, Klein, T, Breed, J, Breeze, A, Overman, R, Phillips, C, Norman, R.A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-12-03 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insights Into Fgfr Kinase Isoform Selectivity: Diverse Binding Modes of Azd4547 and Ponatinib in Complex with Fgfr1 and Fgfr4
Structure, 22, 2014
|
|
6H7B
 
 | | Structure of Leishmania PABP1 (domain J) complexed with a peptide containing the PAM2 motif of eIF4E4. | | Descriptor: | HIS-HIS-MET-ASN-PRO-ASN-ALA-THR-GLU-PHE-MET-PRO, Polyadenylate-binding protein | | Authors: | Cameron, A.D, Firczuk, H, dos Santos Rodrigues, F.H, McCarthy, J.E.G. | | Deposit date: | 2018-07-31 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The Leishmania PABP1-eIF4E4 interface: a novel 5'-3' interaction architecture for trans-spliced mRNAs.
Nucleic Acids Res., 47, 2019
|
|
6H7A
 
 | |
5EW8
 
 | | FIBROBLAST GROWTH FACTOR RECEPTOR 1 IN COMPLEX WITH JNJ-4275693 | | Descriptor: | Fibroblast growth factor receptor 1, SULFATE ION, ~{N}'-(3,5-dimethoxyphenyl)-~{N}'-[3-(1-methylpyrazol-4-yl)quinoxalin-6-yl]-~{N}-propan-2-yl-ethane-1,2-diamine | | Authors: | Ogg, D, Breed, J. | | Deposit date: | 2015-11-20 | | Release date: | 2016-03-30 | | Last modified: | 2016-08-10 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Landscape of activating cancer mutations in FGFR kinases and their differential responses to inhibitors in clinical use.
Oncotarget, 7, 2016
|
|
5FLF
 
 | | DISEASE LINKED MUTATION IN FGFR | | Descriptor: | ACETATE ION, CHLORIDE ION, FIBROBLAST GROWTH FACTOR RECEPTOR 1, ... | | Authors: | Thiyagarajan, N, Bunney, T.D, Katan, M. | | Deposit date: | 2015-10-26 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Landscape of Activating Cancer Mutations in Fgfr Kinases and Their Differential Responses to Inhibitors in Clinical Use.
Oncotarget, 7, 2016
|
|
4AJ4
 
 | | rat LDHA in complex with 4-((2-allylsulfanyl-1,3-benzothizol-6-yl) amino)-4-oxo-butanoic acid | | Descriptor: | 4-oxo-4-{[2-(prop-2-en-1-ylsulfanyl)-1,3-benzothiazol-6-yl]amino}butanoic acid, L-LACTATE DEHYDROGENASE A CHAIN, MALONATE ION | | Authors: | Tucker, J.A, Brassington, C, Hassall, G, Vogtherr, M, Ward, R, Tart, J, Davies, G. | | Deposit date: | 2012-02-15 | | Release date: | 2012-03-21 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Design and Synthesis of Novel Lactate Dehydrogenase a Inhibitors by Fragment-Based Lead Generation
J.Med.Chem., 55, 2012
|
|
