4WIQ
 
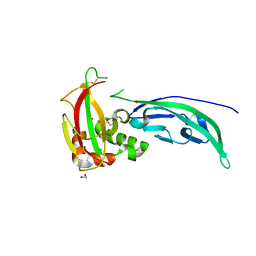 | | The structure of Murine alpha-Dystroglycan T190M mutant N-terminal domain. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Dystroglycan | | Authors: | Brancaccio, A, Lamba, D, Cassetta, A, Bozzi, M, Covaceuszach, S, Sciandra, F, Bigotti, M.G. | | Deposit date: | 2014-09-26 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The Structure of the T190M Mutant of Murine alpha-Dystroglycan at High Resolution: Insight into the Molecular Basis of a Primary Dystroglycanopathy.
Plos One, 10, 2015
|
|
1FCS
 
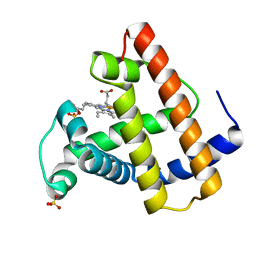 | | CRYSTAL STRUCTURE OF A DISTAL SITE DOUBLE MUTANT OF SPERM WHALE MYOGLOBIN AT 1.6 ANGSTROMS RESOLUTION | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Rizzi, M, Bolognesi, M, Coda, A, Cutruzzola, F, Travaglini Allocatelli, C, Brancaccio, A, Brunori, M. | | Deposit date: | 1993-04-20 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a distal site double mutant of sperm whale myoglobin at 1.6 A resolution.
FEBS Lett., 320, 1993
|
|
1U2C
 
 | |
1FF5
 
 | | STRUCTURE OF E-CADHERIN DOUBLE DOMAIN | | Descriptor: | CALCIUM ION, EPITHELIAL CADHERIN | | Authors: | Pertz, O, Bozic, D, Koch, A.W, Fauser, C, Brancaccio, A, Engel, J. | | Deposit date: | 2000-07-25 | | Release date: | 2000-08-23 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | A new crystal structure, Ca2+ dependence and mutational analysis reveal molecular details of E-cadherin homoassociation.
EMBO J., 18, 1999
|
|
5LLK
 
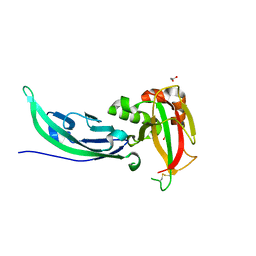 | | Crystal structure of human alpha-dystroglycan | | Descriptor: | 1,2-ETHANEDIOL, Dystroglycan | | Authors: | Covaceuszach, S, Cassetta, A, Lamba, D, Brancaccio, A, Bozzi, M, Sciandra, F, Bigotti, M.G, Konarev, P.V. | | Deposit date: | 2016-07-27 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural flexibility of human alpha-dystroglycan.
FEBS Open Bio, 7, 2017
|
|
5N30
 
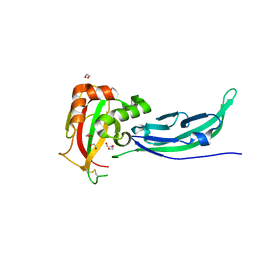 | | Crystal structure of the V72I mutant of the mouse alpha-Dystroglycan N-terminal region | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Dystroglycan | | Authors: | Cassetta, A, Covaceuszach, S, Brancaccio, A, Sciandra, F, Bozzi, M, Bigotti, M.G, Konarev, P.V. | | Deposit date: | 2017-02-08 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The effect of the pathological V72I, D109N and T190M missense mutations on the molecular structure of alpha-dystroglycan.
PLoS ONE, 12, 2017
|
|
5N4H
 
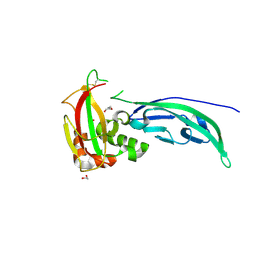 | | Crystal structure of the D109N mutant of the mouse alpha-Dystroglycan N-terminal region | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Dystroglycan | | Authors: | Cassetta, A, Covaceuszach, S, Brancaccio, A, Sciandra, F, Bozzi, M, Bigotti, M.G, Konarev, P.V. | | Deposit date: | 2017-02-10 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | The effect of the pathological V72I, D109N and T190M missense mutations on the molecular structure of alpha-dystroglycan.
PLoS ONE, 12, 2017
|
|
1MNH
 
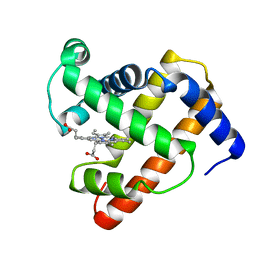 | | INTERACTIONS AMONG RESIDUES CD3, E7, E10 AND E11 IN MYOGLOBINS: ATTEMPTS TO SIMULATE THE O2 AND CO BINDING PROPERTIES OF APLYSIA MYOGLOBIN | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Davies, G.J, Wilkinson, A.J. | | Deposit date: | 1995-01-11 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Interactions among residues CD3, E7, E10, and E11 in myoglobins: attempts to simulate the ligand-binding properties of Aplysia myoglobin.
Biochemistry, 34, 1995
|
|
1MNK
 
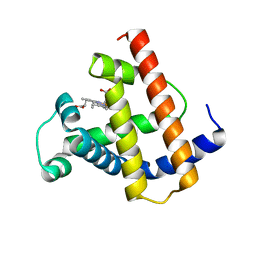 | | INTERACTIONS AMONG RESIDUES CD3, E7, E10 AND E11 IN MYOGLOBINS: ATTEMPTS TO SIMULATE THE O2 AND CO BINDING PROPERTIES OF APLYSIA MYOGLOBIN | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Krzywda, S, Wilkinson, A.J. | | Deposit date: | 1995-01-11 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interactions among residues CD3, E7, E10, and E11 in myoglobins: attempts to simulate the ligand-binding properties of Aplysia myoglobin.
Biochemistry, 34, 1995
|
|
1MNJ
 
 | | INTERACTIONS AMONG RESIDUES CD3, E7, E10 AND E11 IN MYOGLOBINS: ATTEMPTS TO SIMULATE THE O2 AND CO BINDING PROPERTIES OF APLYSIA MYOGLOBIN | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Krzywda, S, Wilkinson, A.J. | | Deposit date: | 1995-01-11 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interactions among residues CD3, E7, E10, and E11 in myoglobins: attempts to simulate the ligand-binding properties of Aplysia myoglobin.
Biochemistry, 34, 1995
|
|
