2BJB
 
 | | Mycobacterium Tuberculosis Epsp Synthase In Unliganded State | | Descriptor: | 3-PHOSPHOSHIKIMATE 1-CARBOXYVINYLTRANSFERASE, ACETATE ION, SODIUM ION | | Authors: | Bourenkov, G.P, Kachalova, G.S, Strizhov, N, Bruning, M, Vagin, A, Bartunik, H.D. | | Deposit date: | 2005-02-01 | | Release date: | 2006-03-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mycobacterium Tuberculosis Epsp Synthase in Unliganded State
To be Published
|
|
2JA2
 
 | | Mycobacterium tuberculosis glutamyl-tRNA synthetase | | Descriptor: | ACETATE ION, CHLORIDE ION, GLUTAMYL-TRNA SYNTHETASE | | Authors: | Bourenkov, G.P, Strizhov, N, Shkolnaya, L.A, Bruning, M, Bartunik, H.D. | | Deposit date: | 2006-11-20 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for Discriminating or Non-Discriminating Functioning of Glutamyl-tRNA Synthetases
To be Published
|
|
3K7M
 
 | | Crystal structure of 6-hydroxy-L-nicotine oxidase from Arthrobacter nicotinovorans | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, 6-hydroxy-L-nicotine oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Bourenkov, G.P, Kachalova, G.S, Bartunik, H.D. | | Deposit date: | 2009-10-13 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure Analysis of Free and Substrate-Bound 6-Hydroxy-l-Nicotine Oxidase from Arthrobacter nicotinovorans.
J.Mol.Biol., 396, 2010
|
|
3K7T
 
 | | Crystal structure of apo-form 6-hydroxy-L-nicotine oxidase, crystal form P3121 | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, 6-hydroxy-L-nicotine oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Bourenkov, G.P, Kachalova, G.S, Bartunik, H.D. | | Deposit date: | 2009-10-13 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure Analysis of Free and Substrate-Bound 6-Hydroxy-l-Nicotine Oxidase from Arthrobacter nicotinovorans.
J.Mol.Biol., 396, 2010
|
|
3K7Q
 
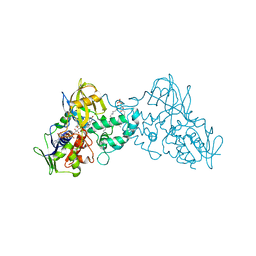 | | Crystal structure of substrate-bound 6-hydroxy-L-nicotine oxidase from Arthrobacter nicotinovorans | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, 5-[(2S)-1-methylpyrrolidin-2-yl]pyridin-2-ol, 6-hydroxy-L-nicotine oxidase, ... | | Authors: | Bourenkov, G.P, Kachalova, G.S, Bartunik, H.D. | | Deposit date: | 2009-10-13 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure Analysis of Free and Substrate-Bound 6-Hydroxy-l-Nicotine Oxidase from Arthrobacter nicotinovorans.
J.Mol.Biol., 396, 2010
|
|
2ZO5
 
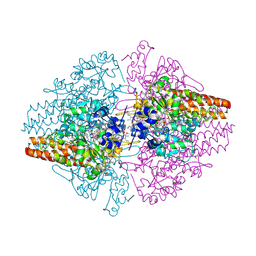 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in a complex with azide | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AZIDE ION, ... | | Authors: | Polyakov, K.M, Boyko, K.M, Slutsky, A, Tikhonova, T.V, Antipov, A.N, Zvyagilskaya, R.A, Popov, A.N, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2008-05-05 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution structural analysis of a novel octaheme cytochrome c nitrite reductase from the haloalkaliphilic bacterium Thioalkalivibrio nitratireducens
J.Mol.Biol., 389, 2009
|
|
1C52
 
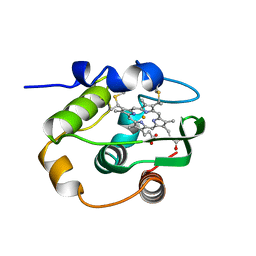 | | THERMUS THERMOPHILUS CYTOCHROME-C552: A NEW HIGHLY THERMOSTABLE CYTOCHROME-C STRUCTURE OBTAINED BY MAD PHASING | | Descriptor: | CYTOCHROME-C552, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Than, M.E, Hof, P, Huber, R, Bourenkov, G.P, Bartunik, H.D, Buse, G, Soulimane, T. | | Deposit date: | 1997-06-23 | | Release date: | 1998-06-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Thermus thermophilus cytochrome-c552: A new highly thermostable cytochrome-c structure obtained by MAD phasing.
J.Mol.Biol., 271, 1997
|
|
1CZA
 
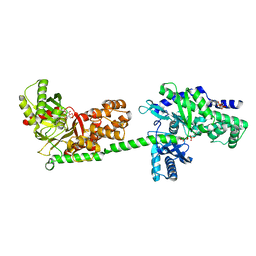 | | MUTANT MONOMER OF RECOMBINANT HUMAN HEXOKINASE TYPE I COMPLEXED WITH GLUCOSE, GLUCOSE-6-PHOSPHATE, AND ADP | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, HEXOKINASE TYPE I, ... | | Authors: | Aleshin, A.E, Liu, X, Kirby, C, Bourenkov, G.P, Bartunik, H.D, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 1999-09-01 | | Release date: | 2000-03-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of mutant monomeric hexokinase I reveal multiple ADP binding sites and conformational changes relevant to allosteric regulation.
J.Mol.Biol., 296, 2000
|
|
3D1I
 
 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in a complex with nitrite | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CALCIUM ION, Eight-heme nitrite reductase, ... | | Authors: | Polyakov, K.M, Boyko, K.M, Slutsky, A, Tikhonova, T.V, Antipov, A.N, Zvyagilskaya, R.A, Popov, A.N, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2008-05-06 | | Release date: | 2009-05-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structural analysis of a novel octaheme cytochrome c nitrite reductase from the haloalkaliphilic bacterium Thioalkalivibrio nitratireducens
J.Mol.Biol., 389, 2009
|
|
2OT4
 
 | | Structure of a hexameric multiheme c nitrite reductase from the extremophile bacterium Thiolkalivibrio nitratireducens | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Polyakov, K.M, Boyko, K.M, Slutsky, A, Tikhonova, T.V, Antipov, A.N, Zvyagilskaya, R.A, Popov, A.N, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2007-02-07 | | Release date: | 2008-04-08 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution structural analysis of a novel octaheme cytochrome c nitrite reductase from the haloalkaliphilic bacterium Thioalkalivibrio nitratireducens.
J.Mol.Biol., 389, 2009
|
|
1D7M
 
 | | COILED-COIL DIMERIZATION DOMAIN FROM CORTEXILLIN I | | Descriptor: | CORTEXILLIN I | | Authors: | Burkhard, P, Kammerer, R.A, Steinmetz, M.O, Bourenkov, G.P, Aebi, U. | | Deposit date: | 1999-10-19 | | Release date: | 2000-03-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The coiled-coil trigger site of the rod domain of cortexillin I unveils a distinct network of interhelical and intrahelical salt bridges.
Structure Fold.Des., 8, 2000
|
|
1DO0
 
 | | ORTHORHOMBIC CRYSTAL FORM OF HEAT SHOCK LOCUS U (HSLU) FROM ESCHERICHIA COLI | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PROTEIN (HEAT SHOCK LOCUS U), ... | | Authors: | Bochtler, M, Hartmann, C, Song, H.K, Bourenkov, G.P, Bartunik, H.D. | | Deposit date: | 1999-12-18 | | Release date: | 2000-02-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structures of HsIU and the ATP-dependent protease HsIU-HsIV.
Nature, 403, 2000
|
|
1DD4
 
 | | Crystal structure of ribosomal protein l12 from thermotoga maritim | | Descriptor: | 50S RIBOSOMAL PROTEIN L7/L12, HEXATANTALUM DODECABROMIDE | | Authors: | Wahl, M.C, Bourenkov, G.P, Bartunik, H.D, Huber, R. | | Deposit date: | 1999-11-08 | | Release date: | 2000-11-13 | | Last modified: | 2023-02-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Flexibility, conformational diversity and two dimerization modes in complexes of ribosomal protein L12.
Embo J., 19, 2000
|
|
1DGK
 
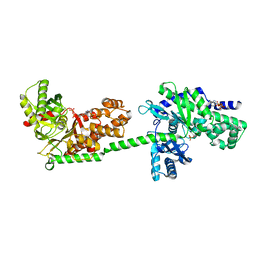 | | MUTANT MONOMER OF RECOMBINANT HUMAN HEXOKINASE TYPE I WITH GLUCOSE AND ADP IN THE ACTIVE SITE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEXOKINASE TYPE I, PHOSPHATE ION, ... | | Authors: | Aleshin, A.E, Liu, X, Kirby, C, Bourenkov, G.P, Bartunik, H.D, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 1999-11-24 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of mutant monomeric hexokinase I reveal multiple ADP binding sites and conformational changes relevant to allosteric regulation.
J.Mol.Biol., 296, 2000
|
|
1DD3
 
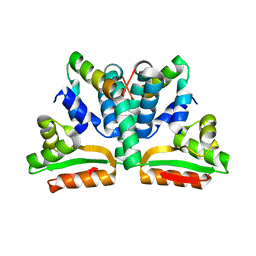 | | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN L12 FROM THERMOTOGA MARITIMA | | Descriptor: | 50S RIBOSOMAL PROTEIN L7/L12 | | Authors: | Wahl, M.C, Bourenkov, G.P, Bartunik, H.D, Huber, R. | | Deposit date: | 1999-11-08 | | Release date: | 2000-11-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Flexibility, conformational diversity and two dimerization modes in complexes of ribosomal protein L12.
EMBO J., 19, 2000
|
|
1KL7
 
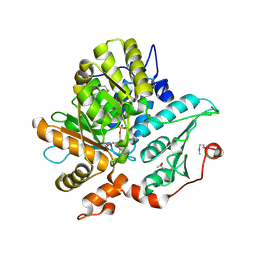 | | Crystal Structure of Threonine Synthase from Yeast | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Threonine Synthase | | Authors: | Garrido-Franco, M, Ehlert, S, Messerschmidt, A, Marinkovic, S, Huber, R, Laber, B, Bourenkov, G.P, Clausen, T. | | Deposit date: | 2001-12-11 | | Release date: | 2002-04-24 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and function of threonine synthase from yeast.
J.Biol.Chem., 277, 2002
|
|
1LI1
 
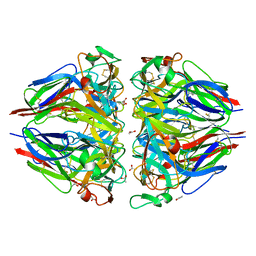 | | The 1.9-A crystal structure of the noncollagenous (NC1) domain of human placenta collagen IV shows stabilization via a novel type of covalent Met-Lys cross-link | | Descriptor: | ACETATE ION, Collagen alpha 1(IV), Collagen alpha 2(IV) | | Authors: | Than, M.E, Henrich, S, Huber, R, Ries, A, Mann, K, Kuhn, K, Timpl, R, Bourenkov, G.P, Bartunik, H.D, Bode, W. | | Deposit date: | 2002-04-17 | | Release date: | 2002-05-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9-A crystal structure of the noncollagenous (NC1) domain of human placenta collagen IV shows stabilization via a novel type of covalent Met-Lys cross-link.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
4DP1
 
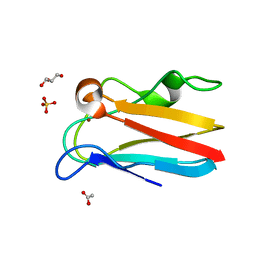 | | The 1.35 Angstrom crystal structure of reduced (CuI) poplar plastocyanin B at pH 4.0 | | Descriptor: | ACETATE ION, COPPER (I) ION, GLYCEROL, ... | | Authors: | Kachalova, G.S, Shosheva, A.H, Bourenkov, G.P, Donchev, A.A, Dimitrov, M.I, Bartunik, H.D. | | Deposit date: | 2012-02-13 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural comparison of the poplar plastocyanin isoforms PCa and PCb sheds new light on the role of the copper site geometry in interactions with redox partners in oxygenic photosynthesis.
J.Inorg.Biochem., 115, 2012
|
|
4DP7
 
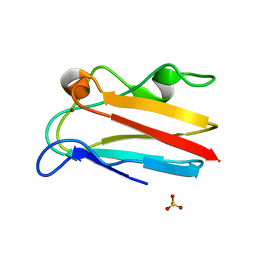 | | The 1.08 Angstrom crystal structure of oxidized (CuII) poplar plastocyanin A at pH 4.0 | | Descriptor: | COPPER (II) ION, Plastocyanin A, chloroplastic, ... | | Authors: | Kachalova, G.S, Shosheva, A.H, Bourenkov, G.P, Donchev, A.A, Dimitrov, M.I, Bartunik, H.D. | | Deposit date: | 2012-02-13 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Structural comparison of the poplar plastocyanin isoforms PCa and PCb sheds new light on the role of the copper site geometry in interactions with redox partners in oxygenic photosynthesis.
J.Inorg.Biochem., 115, 2012
|
|
4DP4
 
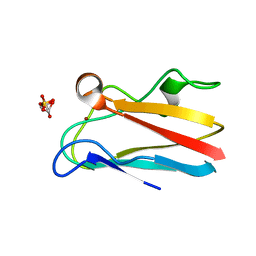 | | The 1.54 Angstrom crystal structure of reduced (CuI) poplar plastocyanin B at pH 6.0 | | Descriptor: | COPPER (I) ION, GLYCEROL, Plastocyanin B, ... | | Authors: | Kachalova, G.S, Shosheva, A.H, Bourenkov, G.P, Donchev, A.A, Dimitrov, M.I, Bartunik, H.D. | | Deposit date: | 2012-02-13 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural comparison of the poplar plastocyanin isoforms PCa and PCb sheds new light on the role of the copper site geometry in interactions with redox partners in oxygenic photosynthesis.
J.Inorg.Biochem., 115, 2012
|
|
4DP6
 
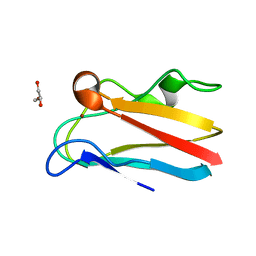 | | The 1.67 Angstrom crystal structure of reduced (CuI) poplar plastocyanin B at pH 8.0 | | Descriptor: | COPPER (I) ION, GLYCEROL, Plastocyanin B, ... | | Authors: | Kachalova, G.S, Shosheva, A.H, Bourenkov, G.P, Donchev, A.A, Dimitrov, M.I, Bartunik, H.D. | | Deposit date: | 2012-02-13 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural comparison of the poplar plastocyanin isoforms PCa and PCb sheds new light on the role of the copper site geometry in interactions with redox partners in oxygenic photosynthesis.
J.Inorg.Biochem., 115, 2012
|
|
4DPC
 
 | | The 1.06 Angstrom crystal structure of reduced (CuI) poplar plastocyanin A at pH 8.0 | | Descriptor: | COPPER (I) ION, Plastocyanin A, chloroplastic | | Authors: | Kachalova, G.S, Shosheva, A.H, Bourenkov, G.P, Donchev, A.A, Dimitrov, M.I, Bartunik, H.D. | | Deposit date: | 2012-02-13 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Structural comparison of the poplar plastocyanin isoforms PCa and PCb sheds new light on the role of the copper site geometry in interactions with redox partners in oxygenic photosynthesis.
J.Inorg.Biochem., 115, 2012
|
|
4DPB
 
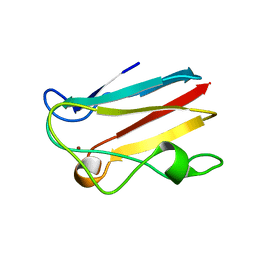 | | The 1.00 Angstrom crystal structure of oxidized (CuII) poplar plastocyanin A at pH 8.0 | | Descriptor: | COPPER (II) ION, Plastocyanin A, chloroplastic | | Authors: | Kachalova, G.S, Shosheva, A.H, Bourenkov, G.P, Donchev, A.A, Dimitrov, M.I, Bartunik, H.D. | | Deposit date: | 2012-02-13 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural comparison of the poplar plastocyanin isoforms PCa and PCb sheds new light on the role of the copper site geometry in interactions with redox partners in oxygenic photosynthesis.
J.Inorg.Biochem., 115, 2012
|
|
4DP2
 
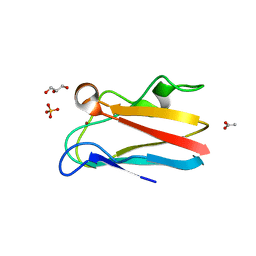 | | The 1.8 Angstrom crystal structure of oxidized (CuII) poplar plastocyanin B at pH 6.0 | | Descriptor: | ACETATE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Kachalova, G.S, Shosheva, A.H, Bourenkov, G.P, Donchev, A.A, Dimitrov, M.I, Bartunik, H.D. | | Deposit date: | 2012-02-13 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural comparison of the poplar plastocyanin isoforms PCa and PCb sheds new light on the role of the copper site geometry in interactions with redox partners in oxygenic photosynthesis.
J.Inorg.Biochem., 115, 2012
|
|
4DP9
 
 | | The 1.00 Angstrom crystal structure of oxidized (CuII) poplar plastocyanin A at pH 6.0 | | Descriptor: | COPPER (II) ION, Plastocyanin A, chloroplastic | | Authors: | Kachalova, G.S, Shosheva, A.H, Bourenkov, G.P, Donchev, A.A, Dimitrov, M.I, Bartunik, H.D. | | Deposit date: | 2012-02-13 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural comparison of the poplar plastocyanin isoforms PCa and PCb sheds new light on the role of the copper site geometry in interactions with redox partners in oxygenic photosynthesis.
J.Inorg.Biochem., 115, 2012
|
|
