4UWM
 
 | | Type II Baeyer-Villiger monooxygenase.The oxygenating constituent of 3,6-diketocamphane monooxygenase from CAM plasmid of Pseudomonas putida in complex with FMN. | | Descriptor: | 3,6-DIKETOCAMPHANE 1,6 MONOOXYGENASE, DI(HYDROXYETHYL)ETHER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Isupov, M.N, Schroeder, E, Gibson, R.P, Beecher, J, Donadio, G, Saneei, V, Dcunha, S, McGhie, E.J, Sayer, C, Davenport, C.F, Lau, P.C, Hasegawa, Y, Iwaki, H, Kadow, M, Loschinski, K, Bornscheuer, U.T, Bourenkov, G, Littlechild, J.A. | | Deposit date: | 2014-08-12 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Oxygenating Constituent of 3,6-Diketocamphane Monooxygenase from the Cam Plasmid of Pseudomonas Putida: The First Crystal Structure of a Type II Baeyer-Villiger Monooxygenase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6EWY
 
 | | RipA Peptidoglycan hydrolase (Rv1477, Mycobacterium tuberculosis) N-terminal domain | | Descriptor: | Peptidoglycan endopeptidase RipA | | Authors: | Schnell, R, Steiner, E.M, Schneider, G, Guy, J, Bourenkov, G. | | Deposit date: | 2017-11-07 | | Release date: | 2018-05-02 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of the N-terminal module of the cell wall hydrolase RipA and its role in regulating catalytic activity.
Proteins, 86, 2018
|
|
4RJI
 
 | | Acetolactate synthase from Bacillus subtilis bound to ThDP - crystal form I | | Descriptor: | Acetolactate synthase, MAGNESIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Sommer, B, von Moeller, H, Haack, M, Qoura, F, Langner, C, Bourenkov, G, Garbe, D, Brueck, T, Loll, B. | | Deposit date: | 2014-10-09 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Detailed Structure-Function Correlations of Bacillus subtilis Acetolactate Synthase.
Chembiochem, 16, 2015
|
|
4RJK
 
 | | Acetolactate synthase from Bacillus subtilis bound to LThDP - crystal form II | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-(1-CARBOXY-1-HYDROXYETHYL)-5-(2-{[HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, Acetolactate synthase, MAGNESIUM ION, ... | | Authors: | Sommer, B, von Moeller, H, Haack, M, Qoura, F, Langner, C, Bourenkov, G, Garbe, D, Brueck, T, Loll, B. | | Deposit date: | 2014-10-09 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Detailed Structure-Function Correlations of Bacillus subtilis Acetolactate Synthase.
Chembiochem, 16, 2015
|
|
4RJJ
 
 | | Acetolactate synthase from Bacillus subtilis bound to ThDP - crystal form II | | Descriptor: | ACETATE ION, Acetolactate synthase, MAGNESIUM ION, ... | | Authors: | Sommer, B, von Moeller, H, Haack, M, Qoura, F, Langner, C, Bourenkov, G, Garbe, D, Brueck, T, Loll, B. | | Deposit date: | 2014-10-09 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Detailed Structure-Function Correlations of Bacillus subtilis Acetolactate Synthase.
Chembiochem, 16, 2015
|
|
1JNR
 
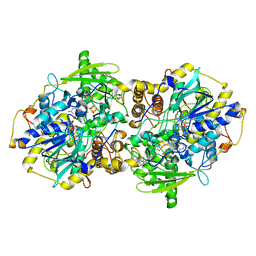 | | Structure of adenylylsulfate reductase from the hyperthermophilic Archaeoglobus fulgidus at 1.6 resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Fritz, G, Roth, A, Schiffer, A, Buechert, T, Bourenkov, G, Bartunik, H.D, Huber, H, Stetter, K.O, Kroneck, P.M.H, Ermler, U. | | Deposit date: | 2001-07-25 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of adenylylsulfate reductase from the hyperthermophilic Archaeoglobus fulgidus at 1.6-A resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JNZ
 
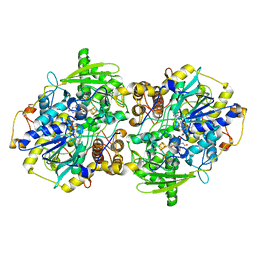 | | Structure of adenylylsulfate reductase from the hyperthermophilic Archaeoglobus fulgidus at 1.6 resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, SULFITE ION, ... | | Authors: | Fritz, G, Roth, A, Schiffer, A, Buechert, T, Bourenkov, G, Bartunik, H.D, Huber, H, Stetter, K.O, Kroneck, P.M, Ermler, U. | | Deposit date: | 2001-07-26 | | Release date: | 2002-03-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of adenylylsulfate reductase from the hyperthermophilic Archaeoglobus fulgidus at 1.6-A resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2AYI
 
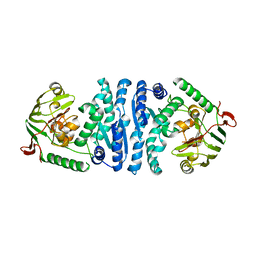 | | Wild-type AmpT from Thermus thermophilus | | Descriptor: | Aminopeptidase T, ZINC ION | | Authors: | Odintsov, S.G, Sabala, I, Bourenkov, G, Rybin, V, Bochtler, M. | | Deposit date: | 2005-09-07 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Substrate Access to the Active Sites in Aminopeptidase T, a Representative of a New Metallopeptidase Clan.
J.Mol.Biol., 354, 2005
|
|
6FVE
 
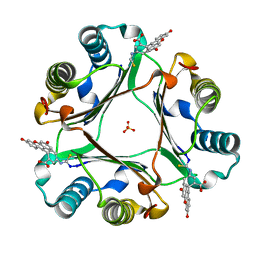 | |
6FVH
 
 | |
4N4Z
 
 | | Trypanosoma brucei procathepsin B structure solved by Serial Microcrystallography using synchrotron radiation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cysteine peptidase C (CPC), beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Gati, C, Bourenkov, G, Klinge, M, Rehders, D, Stellato, F, Oberthuer, D, White, T.A, Yevanov, O, Sommer, B.P, Mogk, S, Duszenko, M, Betzel, C, Schneider, T.R, Chapman, H.N, Redecke, L. | | Deposit date: | 2013-10-08 | | Release date: | 2014-02-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Serial crystallography on in vivo grown microcrystals using synchrotron radiation.
IUCrJ, 1, 2014
|
|
5AEC
 
 | | Type II Baeyer-Villiger monooxygenase.The oxygenating constituent of 3,6-diketocamphane monooxygenase from CAM plasmid of Pseudomonas putida in complex with FMN. | | Descriptor: | 3,6-DIKETOCAMPHANE 1,6 MONOOXYGENASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Isupov, M.N, Schroeder, E, Gibson, R.P, Beecher, J, Donadio, G, Saneei, V, Dcunha, S, McGhie, E.J, Sayer, C, Davenport, C.F, Lau, P.C, Hasegawa, Y, Iwaki, H, Kadow, M, Loschinski, K, Bornscheuer, U.T, Bourenkov, G, Littlechild, J.A. | | Deposit date: | 2015-08-28 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The Oxygenating Constituent of 3,6-Diketocamphane Monooxygenase from the Cam Plasmid of Pseudomonas Putida: The First Crystal Structure of a Type II Baeyer-Villiger Monooxygenase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5APD
 
 | | Hen Egg White Lysozyme not illuminated with 0.4THz radiation | | Descriptor: | LYSOZYME C, SODIUM ION | | Authors: | Lundholm, I, Rodilla, H, Wahlgren, W.Y, Duelli, A, Bourenkov, G, Vukusic, J, Friedman, R, Stake, J, Schneider, T, Katona, G. | | Deposit date: | 2015-09-15 | | Release date: | 2016-01-13 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Terahertz Radiation Induces Non-Thermal Structural Changes Associated with Frohlich Condensation in a Protein Crystal
Struct.Dyn., 2, 2015
|
|
5APE
 
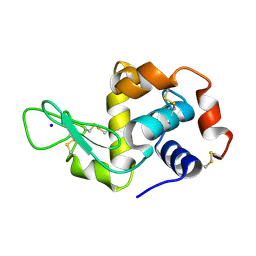 | | Hen Egg White Lysozyme reference dataset odd frames | | Descriptor: | LYSOZYME C, SODIUM ION | | Authors: | Lundholm, I, Rodilla, H, Wahlgren, W.Y, Duelli, A, Bourenkov, G, Vukusic, J, Friedman, R, Stake, J, Schneider, T, Katona, G. | | Deposit date: | 2015-09-15 | | Release date: | 2016-01-13 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Terahertz Radiation Induces Non-Thermal Structural Changes Associated with Frohlich Condensation in a Protein Crystal
Struct.Dyn., 2, 2015
|
|
5APF
 
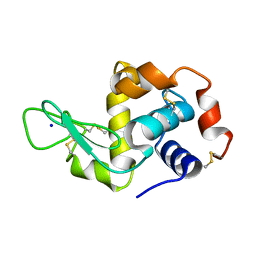 | | Hen Egg White Lysozyme reference dataset even frames | | Descriptor: | LYSOZYME C, SODIUM ION | | Authors: | Lundholm, I, Rodilla, H, Wahlgren, W.Y, Duelli, A, Bourenkov, G, Vukusic, J, Friedman, R, Stake, J, Schneider, T, Katona, G. | | Deposit date: | 2015-09-15 | | Release date: | 2016-01-13 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Terahertz Radiation Induces Non-Thermal Structural Changes Associated with Frohlich Condensation in a Protein Crystal
Struct.Dyn., 2, 2015
|
|
5APC
 
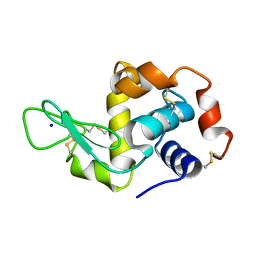 | | Hen Egg White Lysozyme illuminated with 0.4THz radiation | | Descriptor: | LYSOZYME C, SODIUM ION | | Authors: | Lundholm, I, Rodilla, H, Wahlgren, W.Y, Duelli, A, Bourenkov, G, Vukusic, J, Friedman, R, Stake, J, Schneider, T, Katona, G. | | Deposit date: | 2015-09-15 | | Release date: | 2016-01-13 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Terahertz Radiation Induces Non-Thermal Structural Changes Associated with Frohlich Condensation in a Protein Crystal
Struct.Dyn., 2, 2015
|
|
1E6E
 
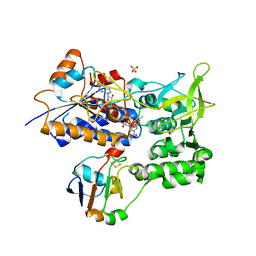 | | ADRENODOXIN REDUCTASE/ADRENODOXIN COMPLEX OF MITOCHONDRIAL P450 SYSTEMS | | Descriptor: | ADRENODOXIN, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Mueller, J.J, Lapko, A, Bourenkov, G, Ruckpaul, K, Heinemann, U. | | Deposit date: | 2000-08-15 | | Release date: | 2001-08-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Adrenodoxin Reductase-Adrenodoxin Complex Structure Suggests Electron Transfer Path in Steroid Biosynthesis.
J.Biol.Chem., 276, 2001
|
|
2V9D
 
 | | Crystal Structure of YagE, a prophage protein belonging to the dihydrodipicolinic acid synthase family from E. coli K12 | | Descriptor: | YAGE | | Authors: | Manicka, S, Peleg, Y, Unger, T, Albeck, S, Dym, O, Greenblatt, H.M, Bourenkov, G, Lamzin, V, Krishnaswamy, S, Sussman, J.L. | | Deposit date: | 2007-08-23 | | Release date: | 2008-03-04 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Yage, a Putative Dhdps Like Protein from Escherichia Coli K12.
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
2V8Z
 
 | | Crystal Structure of YagE, a prophage protein belonging to the dihydrodipicolinic acid synthase family from E. coli K12 | | Descriptor: | YAGE | | Authors: | Manicka, S, Peleg, Y, Unger, T, Albeck, S, Dym, O, Greenblatt, H.M, Bourenkov, G, Lamzin, V, Krishnaswamy, S, Sussman, J.L. | | Deposit date: | 2007-08-16 | | Release date: | 2008-03-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Yage, a Putative Dhdps Like Protein from Escherichia Coli K12.
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
7YXA
 
 | | XFEL crystal structure of the human sphingosine 1 phosphate receptor 5 in complex with ONO-5430608 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[6-(2-naphthalen-1-ylethoxy)-2,3,4,5-tetrahydro-1H-3-benzazepin-3-ium-3-yl]butanoic acid, ... | | Authors: | Lyapina, E, Marin, E, Gusach, A, Orekhov, P, Gerasimov, A, Luginina, A, Vakhrameev, D, Ergasheva, M, Kovaleva, M, Khusainov, G, Khorn, P, Shevtsov, M, Kovalev, K, Okhrimenko, I, Bukhdruker, S, Popov, P, Hu, H, Weierstall, U, Liu, W, Cho, Y, Gushchin, I, Rogachev, A, Bourenkov, G, Park, S, Park, G, Huyn, H.J, Park, J, Gordeliy, V, Borshchevskiy, V, Mishin, A, Cherezov, V. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for receptor selectivity and inverse agonism in S1P 5 receptors.
Nat Commun, 13, 2022
|
|
8C53
 
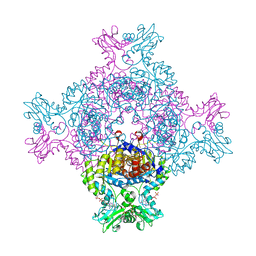 | | Trypanosoma brucei IMP dehydrogenase (ori) crystallized in High Five cells reveals native ligands ATP, GDP and phosphate. Diffraction data collection at 100 K in cellulo; CrystFEL processing | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Lahey-Rudolph, J.M, Schoenherr, R, Boger, J, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Duden, R, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-01-06 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
8C51
 
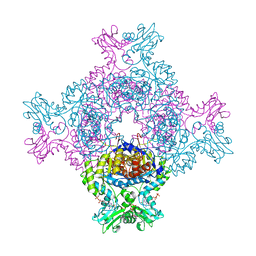 | | Trypanosoma brucei IMP dehydrogenase (cyto) crystallized in High Five cells revealing native ligands ATP, GDP and phosphate. Diffraction data collection at 100 K in cellulo | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Lahey-Rudolph, J.M, Schoenherr, R, Boger, J, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Duden, R, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-01-06 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
8C5K
 
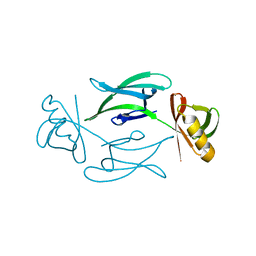 | | HEX-1 (in cellulo, in situ) crystallized and diffracted in High Five cells. Growth and SX data collection at 296 K on CrystalDirect plates | | Descriptor: | Woronin body major protein | | Authors: | Lahey-Rudolph, J.M, Schoenherr, R, Boger, J, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Duden, R, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-01-09 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
8CD5
 
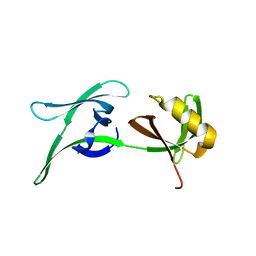 | | structure of HEX-1 from N. crassa crystallized in cellulo, diffracted at 100K and resolved using CrystFEL | | Descriptor: | Woronin body major protein | | Authors: | Boger, J, Schoenherr, R, Lahey-Rudolph, J.M, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-01-30 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
8CD4
 
 | | structure of HEX-1 from N. crassa crystallized in cellulo (cytosol), diffracted at 100K and resolved using CrystFEL | | Descriptor: | Woronin body major protein | | Authors: | Boger, J, Schoenherr, R, Lahey-Rudolph, J.M, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-01-30 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
