1HV2
 
 | | SOLUTION STRUCTURE OF YEAST ELONGIN C IN COMPLEX WITH A VON HIPPEL-LINDAU PEPTIDE | | Descriptor: | ELONGIN C, VON HIPPEL-LINDAU DISEASE TUMOR SUPPRESSOR | | Authors: | Botuyan, M.V, Mer, G, Yi, G.-S, Koth, C.M, Case, D.A, Edwards, A.M, Chazin, W.J, Arrowsmith, C.H. | | Deposit date: | 2001-01-05 | | Release date: | 2001-09-06 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of yeast elongin C in complex with a von Hippel-Lindau peptide.
J.Mol.Biol., 312, 2001
|
|
6CO2
 
 | | Structure of an engineered protein (NUDT16TI) in complex with 53BP1 Tudor domains | | Descriptor: | NUDT16-Tudor-interacting (NUDT16TI), TP53-binding protein 1 | | Authors: | Botuyan, M.V, Thompson, J.R, Cui, G, Mer, G. | | Deposit date: | 2018-03-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Mechanism of 53BP1 activity regulation by RNA-binding TIRR and a designer protein.
Nat. Struct. Mol. Biol., 25, 2018
|
|
1CUR
 
 | | REDUCED RUSTICYANIN, NMR | | Descriptor: | COPPER (II) ION, CU(I) RUSTICYANIN | | Authors: | Botuyan, M.V, Dyson, H.J. | | Deposit date: | 1996-04-19 | | Release date: | 1996-11-08 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of Cu(I) rusticyanin from Thiobacillus ferrooxidans: structural basis for the extreme acid stability and redox potential.
J.Mol.Biol., 263, 1996
|
|
6CO1
 
 | |
6D0L
 
 | | Structure of human TIRR | | Descriptor: | Tudor-interacting repair regulator protein | | Authors: | Cui, G, Botuyan, M.V, Mer, G. | | Deposit date: | 2018-04-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Mechanism of 53BP1 activity regulation by RNA-binding TIRR and a designer protein.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7LIP
 
 | | X-ray structure of SPOP MATH domain (D140G) | | Descriptor: | SULFATE ION, Speckle-type POZ protein | | Authors: | Botuyan, M.V, Cui, G, Mer, G. | | Deposit date: | 2021-01-27 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | ATM-phosphorylated SPOP contributes to 53BP1 exclusion from chromatin during DNA replication.
Sci Adv, 7, 2021
|
|
7LIN
 
 | |
7LIO
 
 | |
7LIQ
 
 | |
2FHD
 
 | | Crystal structure of Crb2 tandem tudor domains | | Descriptor: | DNA repair protein rhp9/CRB2, PHOSPHATE ION | | Authors: | Lee, J, Botuyan, M.V, Thompson, J.R, Mer, G. | | Deposit date: | 2005-12-23 | | Release date: | 2007-01-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair.
Cell(Cambridge,Mass.), 127, 2006
|
|
2G3R
 
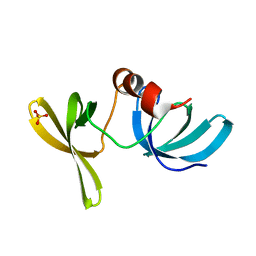 | | Crystal Structure of 53BP1 tandem tudor domains at 1.2 A resolution | | Descriptor: | SULFATE ION, Tumor suppressor p53-binding protein 1 | | Authors: | Lee, J, Botuyan, M.V, Thompson, J.R, Mer, G. | | Deposit date: | 2006-02-20 | | Release date: | 2007-01-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Basis for the Methylation State-Specific Recognition of Histone H4-K20 by 53BP1 and Crb2 in DNA Repair.
Cell(Cambridge,Mass.), 127, 2006
|
|
6VE5
 
 | |
2B02
 
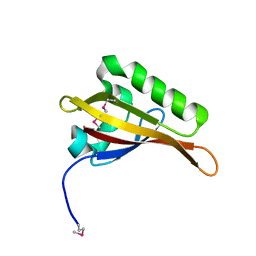 | | Crystal Structure of ARNT PAS-B Domain | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator | | Authors: | Lee, J, Botuyan, M.V, Nomine, Y, Ohh, M, Thompson, J.R, Mer, G. | | Deposit date: | 2005-09-12 | | Release date: | 2006-10-24 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure and Binding Properties of ARNT PAS-B Heterodimerization Domain
To be Published
|
|
3SD4
 
 | |
3P8D
 
 | | Crystal structure of the second Tudor domain of human PHF20 (homodimer form) | | Descriptor: | Medulloblastoma antigen MU-MB-50.72 | | Authors: | Cui, G, Lee, J, Thompson, J.R, Botuyan, M.V, Mer, G. | | Deposit date: | 2010-10-13 | | Release date: | 2011-06-22 | | Last modified: | 2012-09-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | PHF20 is an effector protein of p53 double lysine methylation that stabilizes and activates p53.
Nat.Struct.Mol.Biol., 19, 2012
|
|
6MXY
 
 | |
6MXZ
 
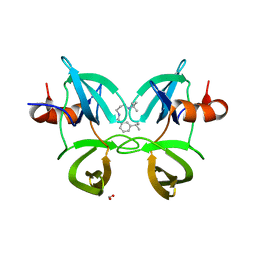 | | Structure of 53BP1 Tudor domains in complex with small molecule UNC3474 | | Descriptor: | FORMIC ACID, N-[3-(tert-butylamino)propyl]-3-(propan-2-yl)benzamide, TP53-binding protein 1 | | Authors: | Cui, G, Botuyan, M.V, Schuller, D.J, Mer, G. | | Deposit date: | 2018-10-31 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An autoinhibited state of 53BP1 revealed by small molecule antagonists and protein engineering.
Nat Commun, 14, 2023
|
|
6MY0
 
 | |
6MXX
 
 | | Structure of 53BP1 tandem Tudor domains in complex with small molecule UNC2991 | | Descriptor: | FORMIC ACID, N-[3-(tert-butylamino)propyl]-3-iodobenzamide, PHOSPHATE ION, ... | | Authors: | Cui, G, Botuyan, M.V, Mer, G. | | Deposit date: | 2018-10-31 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | An autoinhibited state of 53BP1 revealed by small molecule antagonists and protein engineering.
Nat Commun, 14, 2023
|
|
2MWO
 
 | |
2MWP
 
 | |
2QQR
 
 | | JMJD2A hybrid tudor domains | | Descriptor: | JmjC domain-containing histone demethylation protein 3A, SULFATE ION | | Authors: | Lee, J, Botuyan, M.V, Mer, G. | | Deposit date: | 2007-07-26 | | Release date: | 2007-12-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Distinct binding modes specify the recognition of methylated histones H3K4 and H4K20 by JMJD2A-tudor.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2QQS
 
 | |
7KLZ
 
 | |
7LYC
 
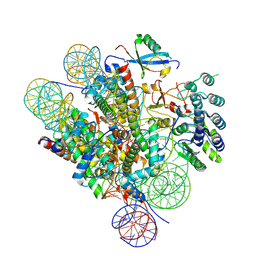 | | Cryo-EM structure of the human nucleosome core particle ubiquitylated at histone H2A Lys13 and Lys15 in complex with BARD1 (residues 415-777) | | Descriptor: | BRCA1-associated RING domain protein 1, DNA (146-MER), DNA (147-MER), ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, D, Mer, E, Mer, G. | | Deposit date: | 2021-03-06 | | Release date: | 2021-06-16 | | Last modified: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Mechanisms of BRCA1-BARD1 nucleosome recognition and ubiquitylation.
Nature, 596, 2021
|
|
