6ZOM
 
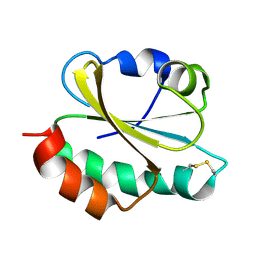 | | Oxidized thioredoxin 1 from the anaerobic bacteria Desulfovibrio vulgaris Hildenborough | | Descriptor: | Thioredoxin | | Authors: | Garcin, E, Bornet, O, Nouailler, M, Pieulle, L, Guerlesquin, F, Sebban-Kreuzer, C. | | Deposit date: | 2020-07-07 | | Release date: | 2021-07-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Glutamate optimizes enzymatic activity under high hydrostatic pressure in Desulfovibrio species: effects on the ubiquitous thioredoxin system.
Extremophiles, 25, 2021
|
|
6T33
 
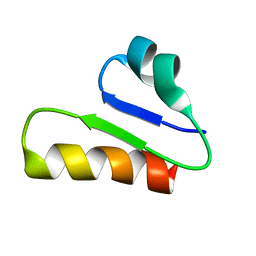 | | The unusual structure of Ruminococcin C1 antimicrobial peptide confers activity against clinical pathogens | | Descriptor: | Ruminococcin C | | Authors: | Chiumento, S, Roblin, C, Bornet, O, Nouailler, M, Muller, C, Basset, C, Kieffer-Jaquinod, S, Coute, Y, Torelli, S, Le Pape, L, Shunemann, V, Jeannot, K, Nicoletti, C, Iranzo, O, Maresca, M, Giardina, T, Fons, M, Devillard, E, Perrier, J, Atta, M, Guerlesquin, F, Lafond, M, Duarte, V. | | Deposit date: | 2019-10-10 | | Release date: | 2020-08-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The unusual structure of Ruminococcin C1 antimicrobial peptide confers clinical properties.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6FW4
 
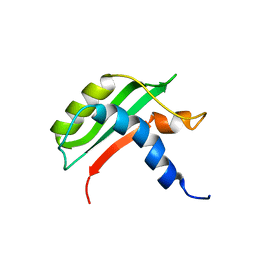 | | Protein-protein interactions and conformational changes : Importance of the hydrophobic cavity of TolA C-terminal domain | | Descriptor: | TolA protein | | Authors: | Navarro, R, van Heijenoort, C, Bornet, O, Houot, L, Lloubes, R, Guerlesquin, F, Nouailler, M. | | Deposit date: | 2018-03-05 | | Release date: | 2019-03-20 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | Induced fit conformational changes of Vibrio cholerae TolAIII domain during the complex formation with the viral PIIIN1 domain: Structural and High-pressure NMR studies.
To Be Published
|
|
2AUV
 
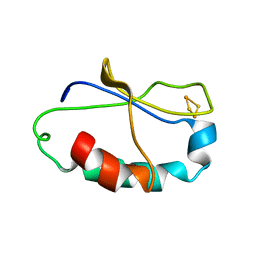 | | Solution Structure of HndAc : A Thioredoxin-like [2Fe-2S] Ferredoxin Involved in the NADP-reducing Hydrogenase Complex | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, potential NAD-reducing hydrogenase subunit | | Authors: | Nouailler, M, Morelli, X, Bornet, O, Chetrit, B, Dermoun, Z, Guerlesquin, F. | | Deposit date: | 2005-08-29 | | Release date: | 2006-06-27 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of HndAc: a thioredoxin-like domain involved in the NADP-reducing hydrogenase complex
Protein Sci., 15, 2006
|
|
2BMT
 
 | | SCORPION TOXIN BMTX2 FROM BUTHUS MARTENSII KARSCH, NMR, 25 STRUCTURES | | Descriptor: | TOXIN BMTX2 | | Authors: | Blanc, E, Romi-Lebrun, R, Bornet, O, Nakajima, T, Darbon, H. | | Deposit date: | 1998-06-16 | | Release date: | 1999-01-13 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Solution structure of two new toxins from the venom of the Chinese scorpion Buthus martensi Karsch blockers of potassium channels.
Biochemistry, 37, 1998
|
|
7Z3C
 
 | | A novel molecular switch controls assembly of bacterial focal adhesions in response to changes in surface structure. | | Descriptor: | Adventurous gliding motility protein GltJ | | Authors: | Attia, B, My, L, Castaing, J.P, Le Guenno, H, Espinosa, L, Schmidt, V, Nouailler, M, Bornet, O, Elantak, L, Mignot, T. | | Deposit date: | 2022-03-02 | | Release date: | 2023-02-22 | | Method: | SOLUTION NMR | | Cite: | A novel molecular switch controls assembly of bacterial focal adhesions
To Be Published
|
|
7ZOK
 
 | | A novel molecular switch controls assembly of bacterial focal adhesions in response to changes in surface structure. | | Descriptor: | Adventurous gliding motility protein GltJ, ZINC ION | | Authors: | Attia, B, My, L, Castaing, J.P, Le Guenno, H, Espinosa, L, Schmidt, V, Nouailler, M, Bornet, O, Mignot, T, Elantak, L. | | Deposit date: | 2022-04-25 | | Release date: | 2023-02-22 | | Method: | SOLUTION NMR | | Cite: | A novel molecular switch controls assembly of bacterial focal adhesions in response to changes in surface structure.
To Be Published
|
|
1BIG
 
 | | SCORPION TOXIN BMTX1 FROM BUTHUS MARTENSII KARSCH, NMR, 25 STRUCTURES | | Descriptor: | TOXIN BMTX1 | | Authors: | Blanc, E, Romi-Lebrun, R, Bornet, O, Nakajima, T, Darbon, H. | | Deposit date: | 1998-06-16 | | Release date: | 1999-01-13 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Solution structure of two new toxins from the venom of the Chinese scorpion Buthus martensi Karsch blockers of potassium channels.
Biochemistry, 37, 1998
|
|
1BKT
 
 | | BMKTX TOXIN FROM SCORPION BUTHUS MARTENSII KARSCH, NMR, 25 STRUCTURES | | Descriptor: | BMKTX | | Authors: | Renisio, J.G, Romi-Lebrun, R, Blanc, E, Bornet, O, Nakajima, T, Darbon, H. | | Deposit date: | 1998-07-03 | | Release date: | 1999-01-13 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmKTX, a K+ blocker toxin from the Chinese scorpion Buthus Martensi
Proteins, 38, 2000
|
|
1E08
 
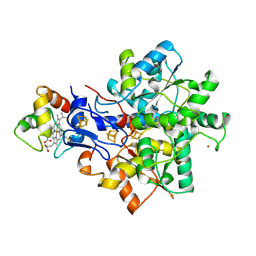 | | Structural model of the [Fe]-Hydrogenase/cytochrome c553 complex combining NMR and soft-docking | | Descriptor: | 1,3-PROPANEDITHIOL, CARBON MONOXIDE, CYANIDE ION, ... | | Authors: | Morelli, X, Czjzek, M, Hatchikian, C.E, Bornet, O, Fontecilla-Camps, J.C, Palma, N.P, Moura, J.J.G, Guerlesquin, F. | | Deposit date: | 2000-03-13 | | Release date: | 2000-08-25 | | Last modified: | 2019-11-27 | | Method: | SOLUTION NMR, THEORETICAL MODEL | | Cite: | Structural Model of the Fe-Hydrogenase/Cytochrome C553 Complex Combining Transverse Relaxation-Optimized Spectroscopy Experiments and Soft Docking Calculations.
J.Biol.Chem., 275, 2000
|
|
1EHX
 
 | | NMR SOLUTION STRUCTURE OF THE LAST UNKNOWN MODULE OF THE CELLULOSOMAL SCAFFOLDIN PROTEIN CIPC OF CLOSTRIDUM CELLULOLYTICUM | | Descriptor: | SCAFFOLDIN PROTEIN | | Authors: | Mosbah, A, Belaich, A, Bornet, O, Belaich, J.P, Henrissat, B, Darbon, H. | | Deposit date: | 2000-02-23 | | Release date: | 2000-11-17 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the module X2 1 of unknown function of the cellulosomal scaffolding protein CipC of Clostridium cellulolyticum.
J.Mol.Biol., 304, 2000
|
|
2L6C
 
 | |
2LKQ
 
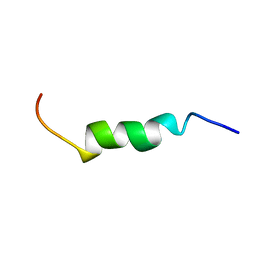 | | NMR structure of the lambda 5 22-45 peptide | | Descriptor: | Immunoglobulin lambda-like polypeptide 1 | | Authors: | Elantak, L, Espeli, M, Boned, A, Bornet, O, Breton, C, Feracci, M, Roche, P, Guerlesquin, F, Schiff, C. | | Deposit date: | 2011-10-19 | | Release date: | 2012-10-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Galectin-1-dependent Pre-B Cell Receptor (Pre-BCR) Activation.
J.Biol.Chem., 287, 2012
|
|
2LRC
 
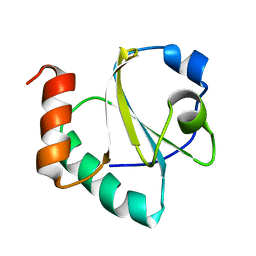 | |
2L6D
 
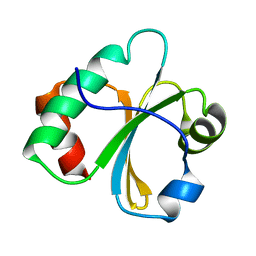 | |
5O7J
 
 | | Structural insights into the periplasmic sensor domain of the GacS histidine kinase controlling biofilm formation in Pseudomonas aeruginosa | | Descriptor: | Histidine kinase | | Authors: | Ali-Ahmad, A, Bornet, O, Fadel, F, Bourne, Y, Vincent, F, Guerlesquin, F, Sebban-Kreuzer, C. | | Deposit date: | 2017-06-09 | | Release date: | 2018-02-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and functional insights into the periplasmic detector domain of the GacS histidine kinase controlling biofilm formation in Pseudomonas aeruginosa.
Sci Rep, 7, 2017
|
|
1G9E
 
 | | SOLUTION STRUCTURE AND RELAXATION MEASUREMENTS OF AN ANTIGEN-FREE HEAVY CHAIN VARIABLE DOMAIN (VHH) FROM LLAMA | | Descriptor: | H14 | | Authors: | Renisio, J.-G, Perez, J, Czisch, M, Guenneugues, M, Bornet, O, Frenken, L, Cambillau, C, Darbon, H. | | Deposit date: | 2000-11-23 | | Release date: | 2002-10-23 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of an antigen-free heavy chain variable
domain (VHH) from Llama
Proteins, 47, 2002
|
|
1GX7
 
 | | Best model of the electron transfer complex between cytochrome c3 and [Fe]-hydrogenase | | Descriptor: | 1,3-PROPANEDITHIOL, CARBON MONOXIDE, CYANIDE ION, ... | | Authors: | Elantak, L, Morelli, X, Bornet, O, Hatchikian, C, Czjzek, M, Dolla, A, Guerlesquin, F. | | Deposit date: | 2002-03-28 | | Release date: | 2003-07-31 | | Last modified: | 2019-11-27 | | Method: | SOLUTION NMR, THEORETICAL MODEL | | Cite: | The Cytochrome C(3)-[Fe]-Hydrogenase Electron-Transfer Complex: Structural Model by NMR Restrained Docking
FEBS Lett., 548, 2003
|
|
1LIR
 
 | | LQ2 FROM LEIURUS QUINQUESTRIATUS, NMR, 22 STRUCTURES | | Descriptor: | LQ2 | | Authors: | Renisio, J.G, Lu, Z, Blanc, E, Jin, W, Lewis, J.H, Bornet, O, Darbon, H. | | Deposit date: | 1998-04-02 | | Release date: | 1998-06-17 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Solution structure of potassium channel-inhibiting scorpion toxin Lq2.
Proteins, 34, 1999
|
|
1VK8
 
 | |
1GMM
 
 | | Carbohydrate binding module CBM6 from xylanase U Clostridium thermocellum | | Descriptor: | CALCIUM ION, CBM6, SODIUM ION, ... | | Authors: | Czjzek, M, Mosbah, A, Bolam, D, Allouch, J, Zamboni, V, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2001-09-19 | | Release date: | 2001-11-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Location of the Ligand-Binding Site of Carbohydrate-Binding Modules that Have Evolved from a Common Sequence is not Conserved.
J.Biol.Chem., 276, 2001
|
|
1K19
 
 | | NMR Solution Structure of the Chemosensory Protein CSP2 from Moth Mamestra brassicae | | Descriptor: | Chemosensory Protein CSP2 | | Authors: | Mosbah, A, Campanacci, V, Lartigue, A, Tegoni, M, Cambillau, C, Darbon, H. | | Deposit date: | 2001-09-24 | | Release date: | 2002-12-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a chemosensory protein from the moth Mamestra brassicae
BIOCHEM.J., 369, 2003
|
|
1KX9
 
 | | ANTENNAL CHEMOSENSORY PROTEIN A6 FROM THE MOTH MAMESTRA BRASSICAE | | Descriptor: | ACETATE ION, CHEMOSENSORY PROTEIN A6 | | Authors: | Lartigue, A, Campanacci, V, Roussel, A, Larsson, A.M, Jones, T.A, Tegoni, M, Cambillau, C. | | Deposit date: | 2002-01-31 | | Release date: | 2002-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray structure and ligand binding study of a moth chemosensory protein
J.Biol.Chem., 277, 2002
|
|
1KX8
 
 | | Antennal Chemosensory Protein A6 from Mamestra brassicae, tetragonal form | | Descriptor: | CHEMOSENSORY PROTEIN A6 | | Authors: | Lartigue, A, Campanacci, V, Roussel, A, Larsson, A.M, Jones, T.A, Tegoni, M, Cambillau, C. | | Deposit date: | 2002-01-31 | | Release date: | 2002-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-Ray Structure and Ligand Binding Study of a Chemosensory Protein
J.Biol.Chem., 277, 2002
|
|
