6SYY
 
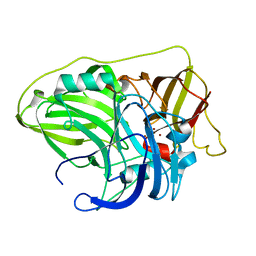 | | Crystal structure of McoA multicopper oxidase from the hyperthermophile Aquifex aeolicus | | Descriptor: | COPPER (II) ION, OXYGEN ATOM, Periplasmic cell division protein (SufI) | | Authors: | Borges, P.T, Brissos, V, Cordeiro, T.N, Martins, L.O, Frazao, C. | | Deposit date: | 2019-10-01 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | The Methionine-Rich Loop of Multicopper Oxidase McoA follows Open-To-Close Transitions with a Role in Enzyme Catalysis
Acs Catalysis, 2020
|
|
6ETB
 
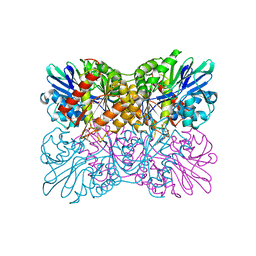 | | Aerobic S262Y mutation of E. coli FLRD core | | Descriptor: | Anaerobic nitric oxide reductase flavorubredoxin, FE (III) ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Borges, P.T, Romao, C.V, Carrondo, M.A, Teixeira, M, Frazao, C. | | Deposit date: | 2017-10-26 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | A tyrosine mutation in E. coli flavodiiron protein increases radiation sensitivity in the crystal structure
To Be Published
|
|
6H0C
 
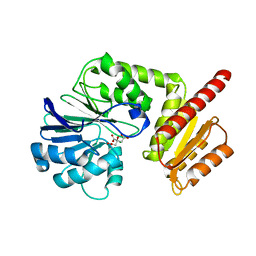 | | Flv1 flavodiiron core from Synechocystis sp. PCC6803 | | Descriptor: | CHLORIDE ION, CITRATE ANION, Putative diflavin flavoprotein A 3 | | Authors: | Borges, P.T, Romao, C.V, Saraiva, L, Goncalves, V.L, Carrondo, M.A, Teixeira, M, Frazao, C. | | Deposit date: | 2018-07-08 | | Release date: | 2019-01-30 | | Last modified: | 2019-02-06 | | Method: | X-RAY DIFFRACTION (1.592 Å) | | Cite: | Analysis of a new flavodiiron core structural arrangement in Flv1-Delta FlR protein from Synechocystis sp. PCC6803.
J. Struct. Biol., 205, 2019
|
|
6H0D
 
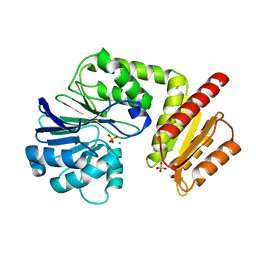 | | Metal soaked Flv1 flavodiiron core from Synechocystis sp. PCC6803 | | Descriptor: | CHLORIDE ION, Putative diflavin flavoprotein A 3, SULFATE ION | | Authors: | Borges, P.T, Romao, C.V, Saraiva, L, Goncalves, V.L, Carrondo, M.A, Teixeira, M, Frazao, C. | | Deposit date: | 2018-07-08 | | Release date: | 2019-01-30 | | Last modified: | 2019-02-06 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Analysis of a new flavodiiron core structural arrangement in Flv1-Delta FlR protein from Synechocystis sp. PCC6803.
J. Struct. Biol., 205, 2019
|
|
4CAB
 
 | | The refined structure of catalase DR1998 from Deinococcus radiodurans at 2.6 A resolution | | Descriptor: | CATALASE, CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Borges, P.T, Miranda, C.S, Santos, S.P, Frazao, C, Romao, C.V. | | Deposit date: | 2013-10-08 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Structure of the Mono-Functional Heme Catalase Dr1998 from Deinococcus Radiodurans
FEBS J., 281, 2014
|
|
7QYZ
 
 | | Crystal structure of a DyP-type peroxidase 6E10 variant from Pseudomonas putida | | Descriptor: | Dyp-type peroxidase family protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Borges, P.T, Silva, D, Martins, L.O, Frazao, C. | | Deposit date: | 2022-01-30 | | Release date: | 2022-08-03 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.452 Å) | | Cite: | Unveiling molecular details behind improved activity at neutral to alkaline pH of an engineered DyP-type peroxidase.
Comput Struct Biotechnol J, 20, 2022
|
|
7QZA
 
 | | Crystal structure of a DyP-type peroxidase 29E4 variant from Pseudomonas putida | | Descriptor: | Dyp-type peroxidase family protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Borges, P.T, Silva, D, Frazao, C, Martins, L.O. | | Deposit date: | 2022-01-30 | | Release date: | 2022-08-03 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Unveiling molecular details behind improved activity at neutral to alkaline pH of an engineered DyP-type peroxidase.
Comput Struct Biotechnol J, 20, 2022
|
|
7QYQ
 
 | | Crystal structure of a DyP-type peroxidase from Pseudomonas putida | | Descriptor: | Dyp-type peroxidase family protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Borges, P.T, Silva, D, Frazao, C, Martins, L.O. | | Deposit date: | 2022-01-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Unveiling molecular details behind improved activity at neutral to alkaline pH of an engineered DyP-type peroxidase.
Comput Struct Biotechnol J, 20, 2022
|
|
7R0F
 
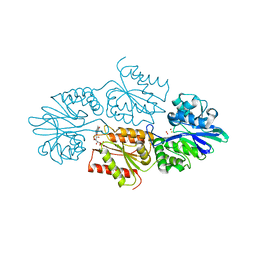 | | Crystal structure of a flavodiiron protein D52K mutant in the oxidized state from Escherichia coli | | Descriptor: | Anaerobic nitric oxide reductase flavorubredoxin, FE (III) ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Borges, P.T, Teixeira, M, Romao, C.V, Frazao, C. | | Deposit date: | 2022-02-02 | | Release date: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Crystal structure of a flavodiiron protein D52K mutant in the oxidized state from Escherichia coli
To Be Published
|
|
7R2P
 
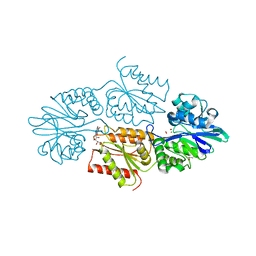 | | Crystal structure of a flavodiiron protein D52K/S262Y mutant in the oxidized state from Escherichia coli | | Descriptor: | Anaerobic nitric oxide reductase flavorubredoxin, FE (III) ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Borges, P.T, Teixeira, M, Romao, C.V, Frazao, C. | | Deposit date: | 2022-02-05 | | Release date: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (2.538 Å) | | Cite: | Crystal structure of a flavodiiron protein D52K/S262Y mutant in the oxidized state from Escherichia coli
To Be Published
|
|
7R2O
 
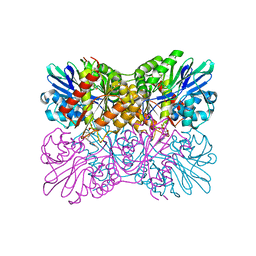 | | Crystal structure of a flavodiiron protein S262Y mutant in the oxidized state from Escherichia coli | | Descriptor: | Anaerobic nitric oxide reductase flavorubredoxin, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Borges, P.T, Teixeira, M, Romao, C.V, Frazao, C. | | Deposit date: | 2022-02-05 | | Release date: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of a flavodiiron protein S262Y mutant in the oxidized state from Escherichia coli
To Be Published
|
|
7R1J
 
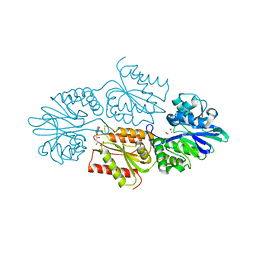 | | Crystal structure of a flavodiiron protein S262Y mutant in the reduced state from Escherichia coli | | Descriptor: | Anaerobic nitric oxide reductase flavorubredoxin, FE (III) ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Borges, P.T, Teixeira, M, Romao, C.V, Frazao, C. | | Deposit date: | 2022-02-03 | | Release date: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Crystal structure of a flavodiiron protein S262Y mutant in the reduced state from Escherichia coli
To Be Published
|
|
7R2S
 
 | | Crystal structure of a flavodiiron protein D52K mutant from Escherichia coli pressurized with krypton gas | | Descriptor: | Anaerobic nitric oxide reductase flavorubredoxin, FE (III) ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Borges, P.T, Teixeira, M, Romao, C.V, Frazao, C. | | Deposit date: | 2022-02-05 | | Release date: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Crystal structure of a flavodiiron protein D52K mutant from Escherichia coli pressurized with krypton gas
To Be Published
|
|
7R1H
 
 | | Crystal structure of a flavodiiron protein D52K mutant in the reduced state from Escherichia coli | | Descriptor: | Anaerobic nitric oxide reductase flavorubredoxin, FE (III) ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Borges, P.T, Teixeira, M, Romao, C.V, Frazao, C. | | Deposit date: | 2022-02-03 | | Release date: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of a flavodiiron protein D52K mutant in the reduced state from Escherichia coli
To Be Published
|
|
7R2R
 
 | | Crystal structure of a flavodiiron protein D52K/S262Y mutant in the reduced state from Escherichia coli | | Descriptor: | Anaerobic nitric oxide reductase flavorubredoxin, FE (III) ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Borges, P.T, Teixeira, M, Romao, C.V, Frazao, C. | | Deposit date: | 2022-02-05 | | Release date: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Crystal structure of a flavodiiron protein D52K/S262Y mutant in the reduced state from Escherichia coli
To Be Published
|
|
8P4G
 
 | | Crystal structure of a multicopper oxidase 3F3 variant from Pyrobaculum aerophilum | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Multicopper oxidase | | Authors: | Borges, P.T, Brissos, V, Frazao, C, Martins, L.O. | | Deposit date: | 2023-05-21 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Flexible active-site loops fine-tune substrate specificity of hyperthermophilic metallo-oxidases.
J.Biol.Inorg.Chem., 2024
|
|
6TTD
 
 | | Crystal structure of McoA multicopper oxidase 2F4 variant from the hyperthermophile Aquifex aeolicus | | Descriptor: | COPPER (II) ION, GLYCEROL, Multicopper oxidase, ... | | Authors: | Borges, P.T, Brissos, V, Cordeiro, T.N, Martins, L.O, Frazao, C. | | Deposit date: | 2019-12-26 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Distal Mutations Shape Substrate-Binding Sites during Evolution of a Metallo-Oxidase into a Laccase
Acs Catalysis, 2022
|
|
8A5C
 
 | |
8A5F
 
 | |
8A5G
 
 | |
7PL0
 
 | | Crystal structure of a DyP-type peroxidase 5G5 variant from Bacillus subtilis | | Descriptor: | Deferrochelatase/peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Borges, P.T, Rodrigues, C, Silva, D, Taborda, A, Brissos, V, Frazao, C, Martins, L.O. | | Deposit date: | 2021-08-27 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Loops around the Heme Pocket Have a Critical Role in the Function and Stability of Bs DyP from Bacillus subtilis .
Int J Mol Sci, 22, 2021
|
|
7PKX
 
 | | Crystal structure of a DyP-type peroxidase from Bacillus subtilis in P3121 space group | | Descriptor: | Deferrochelatase/peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Borges, P.T, Rodrigues, C, Silva, D, Taborda, A, Brissos, V, Frazao, C, Martins, L.O. | | Deposit date: | 2021-08-27 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | Loops around the Heme Pocket Have a Critical Role in the Function and Stability of Bs DyP from Bacillus subtilis .
Int J Mol Sci, 22, 2021
|
|
7QFD
 
 | | Crystal structure of a bacterial pyranose 2-oxidase complex with D-glucose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, GMC oxidoreductase family protein, ... | | Authors: | Borges, P.T, Frazao, T, Taborda, A, Frazao, C, Martins, L.O. | | Deposit date: | 2021-12-05 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanistic insights into glycoside 3-oxidases involved in C-glycoside metabolism in soil microorganisms.
Nat Commun, 14, 2023
|
|
7QF8
 
 | | Crystal structure of a bacterial pyranose 2-oxidase from Pseudoarthrobacter siccitolerans | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, GMC oxidoreductase family protein, ... | | Authors: | Borges, P.T, Frazao, T, Taborda, A, Frazao, C, Martins, L.O. | | Deposit date: | 2021-12-04 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | Mechanistic insights into glycoside 3-oxidases involved in C-glycoside metabolism in soil microorganisms.
Nat Commun, 14, 2023
|
|
7QVA
 
 | | Crystal structure of a bacterial pyranose 2-oxidase in complex with mangiferin | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GMC oxidoreductase family protein, Mangiferin, ... | | Authors: | Borges, P.T, Frazao, T, Taborda, T, Brissos, V, Frazao, C, Martins, L.O. | | Deposit date: | 2022-01-20 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanistic insights into glycoside 3-oxidases involved in C-glycoside metabolism in soil microorganisms.
Nat Commun, 14, 2023
|
|
