2WP8
 
 | | yeast rrp44 nuclease | | Descriptor: | CHLORIDE ION, EXOSOME COMPLEX COMPONENT RRP45, EXOSOME COMPLEX COMPONENT SKI6, ... | | Authors: | Basquin, J, Bonneau, F, Ebert, J, Lorentzen, E, Conti, E. | | Deposit date: | 2009-08-03 | | Release date: | 2009-11-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Yeast Exosome Functions as a Macromolecular Cage to Channel RNA Substrates for Degradation.
Cell(Cambridge,Mass.), 139, 2009
|
|
3N89
 
 | | KH domains | | Descriptor: | Defective in germ line development protein 3, isoform a | | Authors: | Nakel, K, Hartung, S.A, Bonneau, F, Eckmann, C.R, Conti, E. | | Deposit date: | 2010-05-28 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.789 Å) | | Cite: | Four KH domains of the C. elegans Bicaudal-C ortholog GLD-3 form a globular structural platform.
Rna, 16, 2010
|
|
4XR7
 
 | | Structure of the Saccharomyces cerevisiae PAN2-PAN3 core complex | | Descriptor: | PAB-dependent poly(A)-specific ribonuclease subunit PAN2, PAB-dependent poly(A)-specific ribonuclease subunit PAN3 | | Authors: | Schafer, I.B, Rode, M, Bonneau, F, Schussler, S, Conti, E. | | Deposit date: | 2015-01-20 | | Release date: | 2015-01-28 | | Method: | X-RAY DIFFRACTION (3.796 Å) | | Cite: | The structure of the Pan2-Pan3 core complex reveals cross-talk between deadenylase and pseudokinase.
Nat. Struct. Mol. Biol., 21, 2014
|
|
4ZRL
 
 | | Structure of the non canonical Poly(A) polymerase complex GLD-2 - GLD-3 | | Descriptor: | CHLORIDE ION, Defective in germ line development protein 3, Poly(A) RNA polymerase gld-2 | | Authors: | Nakel, K, Bonneau, F, Eckmann, C.R, Conti, E. | | Deposit date: | 2015-05-12 | | Release date: | 2015-07-08 | | Last modified: | 2017-09-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural basis for the activation of the C. elegans noncanonical cytoplasmic poly(A)-polymerase GLD-2 by GLD-3.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6EJ5
 
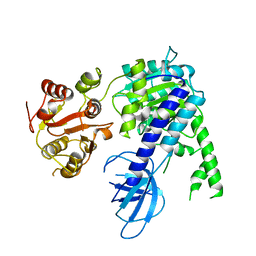 | |
3BXJ
 
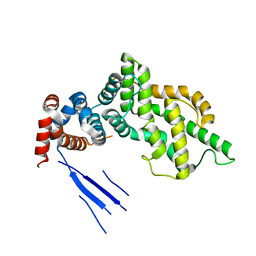 | | Crystal Structure of the C2-GAP Fragment of synGAP | | Descriptor: | Ras GTPase-activating protein SynGAP | | Authors: | Pena, V, Hothorn, M, Eberth, A, Kaschau, N, Parret, A, Gremer, L, Bonneau, F, Ahmadian, M.R, Scheffzek, K. | | Deposit date: | 2008-01-14 | | Release date: | 2008-03-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The C2 domain of SynGAP is essential for stimulation of the Rap GTPase reaction.
Embo Rep., 9, 2008
|
|
7AQO
 
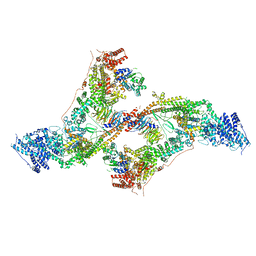 | | yeast THO-Sub2 complex dimer | | Descriptor: | BJ4_G0025130.mRNA.1.CDS.1, EM14S01-3B_G0007820.mRNA.1.CDS.1, TEX1 isoform 1, ... | | Authors: | Schuller, S.K, Schuller, J.M, Prabu, R.J, Baumgartner, M, Bonneau, F, basquin, J, Conti, E. | | Deposit date: | 2020-10-22 | | Release date: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural insights into the nucleic acid remodeling mechanisms of the yeast THO-Sub2 complex.
Elife, 9, 2020
|
|
7APX
 
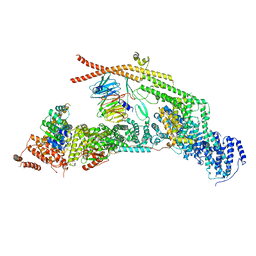 | | yeast THO-Sub2 complex | | Descriptor: | ATP-dependent RNA helicase SUB2, Protein TEX1, THO complex subunit 2,Tho2, ... | | Authors: | Schuller, S.K, Schuller, J.M, Prabu, R.J, Baumgartner, M, Bonneau, F, basquin, J, Conti, E. | | Deposit date: | 2020-10-20 | | Release date: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into the nucleic acid remodeling mechanisms of the yeast THO-Sub2 complex.
Elife, 9, 2020
|
|
4U4C
 
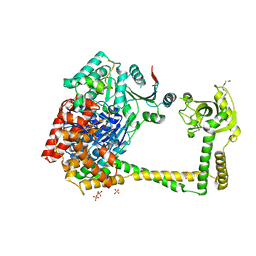 | | The molecular architecture of the TRAMP complex reveals the organization and interplay of its two catalytic activities | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent RNA helicase DOB1, CHLORIDE ION, ... | | Authors: | Falk, S, Weir, J.R, Hentschel, J, Reichelt, P, Bonneau, F, Conti, E. | | Deposit date: | 2014-07-23 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Molecular Architecture of the TRAMP Complex Reveals the Organization and Interplay of Its Two Catalytic Activities.
Mol.Cell, 55, 2014
|
|
4UM2
 
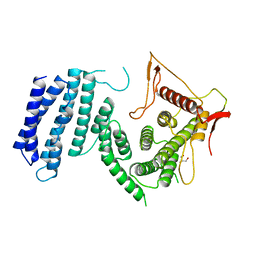 | | Crystal structure of the TPR domain of SMG6 | | Descriptor: | GLYCEROL, TELOMERASE-BINDING PROTEIN EST1A | | Authors: | Chakrabarti, S, Bonneau, F, Schuessler, S, Eppinger, E, Conti, E. | | Deposit date: | 2014-05-14 | | Release date: | 2014-07-23 | | Last modified: | 2014-08-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phospho-Dependent and Phospho-Independent Interactions of the Helicase Upf1 with the Nmd Factors Smg5-Smg7 and Smg6.
Nucleic Acids Res., 42, 2014
|
|
5JNB
 
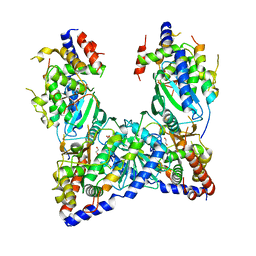 | | structure of GLD-2/RNP-8 complex | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Poly(A) RNA polymerase gld-2, ... | | Authors: | Nakel, K, Bonneau, F, Basquin, C, Eckmann, C.R, Conti, E. | | Deposit date: | 2016-04-29 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.486 Å) | | Cite: | Structural basis for the antagonistic roles of RNP-8 and GLD-3 in GLD-2 poly(A)-polymerase activity.
Rna, 22, 2016
|
|
2XZO
 
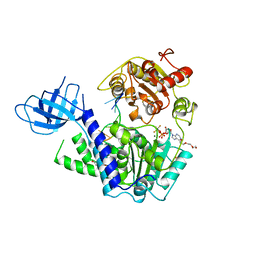 | | Upf1 helicase - RNA complex | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*UP*UP)-3', ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Chakrabarti, S, Jayachandran, U, Bonneau, F, Fiorini, F, Basquin, C, Domcke, S, Le Hir, H, Conti, E. | | Deposit date: | 2010-11-26 | | Release date: | 2011-03-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | Molecular Mechanisms for the RNA-Dependent ATPase Activity of Upf1 and its Regulation by Upf2.
Mol.Cell, 41, 2011
|
|
2XZP
 
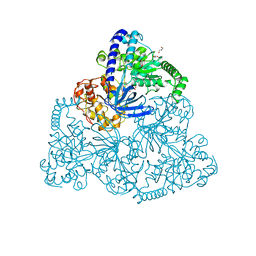 | | Upf1 helicase | | Descriptor: | GLYCEROL, MALONATE ION, REGULATOR OF NONSENSE TRANSCRIPTS 1 | | Authors: | Chakrabarti, S, Jayachandran, U, Bonneau, F, Fiorini, F, Basquin, C, Domcke, S, Le Hir, H, Conti, E. | | Deposit date: | 2010-11-26 | | Release date: | 2011-03-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Molecular Mechanisms for the RNA-Dependent ATPase Activity of Upf1 and its Regulation by Upf2.
Mol.Cell, 41, 2011
|
|
2XZL
 
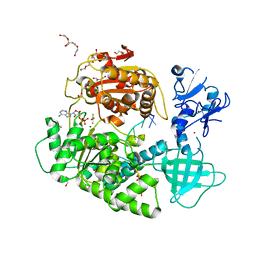 | | Upf1-RNA complex | | Descriptor: | 5- R(*UP*UP*UP*UP*UP*UP*UP*UP*U) -3, ADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT HELICASE NAM7, ... | | Authors: | Chakrabarti, S, Jayachandran, U, Bonneau, F, Fiorini, F, Basquin, C, Domcke, S, Le Hir, H, Conti, E. | | Deposit date: | 2010-11-26 | | Release date: | 2011-03-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Mechanisms for the RNA-Dependent ATPase Activity of Upf1 and its Regulation by Upf2.
Mol.Cell, 41, 2011
|
|
7Z52
 
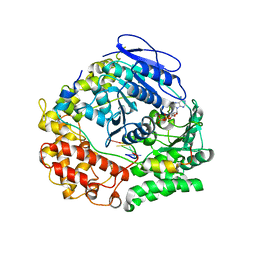 | | Human NEXT dimer - focused reconstruction of the single MTR4 | | Descriptor: | Exosome RNA helicase MTR4, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RNA (5'-R(P*UP*UP*UP*UP*U)-3'), ... | | Authors: | Gerlach, P, Lingaraju, M, Salerno-Kochan, A, Bonneau, F, Basquin, J, Conti, E. | | Deposit date: | 2022-03-07 | | Release date: | 2022-06-22 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and regulation of the nuclear exosome targeting complex guides RNA substrates to the exosome.
Mol.Cell, 82, 2022
|
|
7Z4Y
 
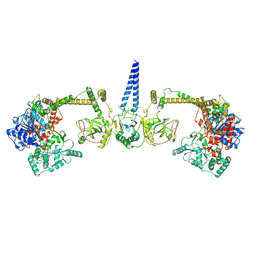 | | Human NEXT dimer - overall reconstruction of the core complex | | Descriptor: | Exosome RNA helicase MTR4, Zinc finger CCHC domain-containing protein 8 | | Authors: | Gerlach, P, Lingaraju, M, Salerno-Kochan, A, Bonneau, F, Basquin, J, Conti, E. | | Deposit date: | 2022-03-06 | | Release date: | 2022-06-22 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure and regulation of the nuclear exosome targeting complex guides RNA substrates to the exosome.
Mol.Cell, 82, 2022
|
|
7Z4Z
 
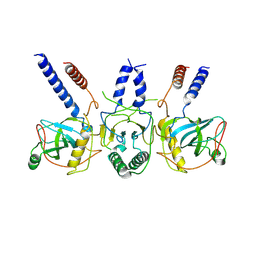 | | Human NEXT dimer - focused reconstruction of the dimerization module | | Descriptor: | Exosome RNA helicase MTR4, Zinc finger CCHC domain-containing protein 8 | | Authors: | Gerlach, P, Lingaraju, M, Salerno-Kochan, A, Bonneau, F, Basquin, J, Conti, E. | | Deposit date: | 2022-03-06 | | Release date: | 2022-06-22 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure and regulation of the nuclear exosome targeting complex guides RNA substrates to the exosome.
Mol.Cell, 82, 2022
|
|
2D4Q
 
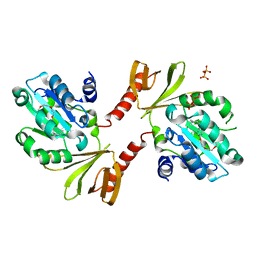 | | Crystal structure of the Sec-PH domain of the human neurofibromatosis type 1 protein | | Descriptor: | Neurofibromin, OXTOXYNOL-10, PYROPHOSPHATE 2- | | Authors: | D'angelo, I, Welti, S, Bonneau, F, Scheffzek, K. | | Deposit date: | 2005-10-22 | | Release date: | 2006-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel bipartite phospholipid-binding module in the neurofibromatosis type 1 protein
Embo Rep., 7, 2006
|
|
2XGJ
 
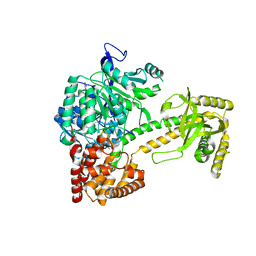 | | Structure of Mtr4, a DExH helicase involved in nuclear RNA processing and surveillance | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT RNA HELICASE DOB1, RNA (5'-(*AP*AP*AP*AP*A)-3') | | Authors: | Weir, J.R, Bonneau, F, Hentschel, J, Conti, E. | | Deposit date: | 2010-06-04 | | Release date: | 2010-06-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Analysis Reveals the Characteristic Features of Mtr4, a Dexh Helicase Involved in Nuclear RNA Processing and Surveillance.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7QE0
 
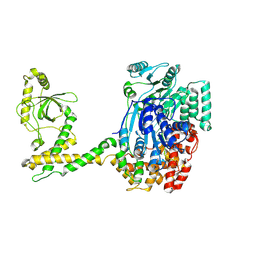 | | 80S-bound human SKI complex in the open state | | Descriptor: | Helicase SKI2W, RNA (5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*U)-3') | | Authors: | Koegel, A, Keidel, A, Bonneau, F, Schaefer, I.B, Conti, E. | | Deposit date: | 2021-12-01 | | Release date: | 2022-02-09 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | The human SKI complex regulates channeling of ribosome-bound RNA to the exosome via an intrinsic gatekeeping mechanism.
Mol.Cell, 82, 2022
|
|
7QDR
 
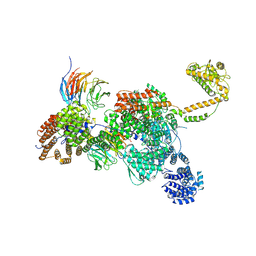 | | Apo human SKI complex in the closed state | | Descriptor: | Helicase SKI2W, Tetratricopeptide repeat protein 37, WD repeat-containing protein 61 | | Authors: | Koegel, A, Keidel, A, Bonneau, F, Schaefer, I.B, Conti, E. | | Deposit date: | 2021-11-29 | | Release date: | 2022-02-02 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The human SKI complex regulates channeling of ribosome-bound RNA to the exosome via an intrinsic gatekeeping mechanism.
Mol.Cell, 82, 2022
|
|
7QDZ
 
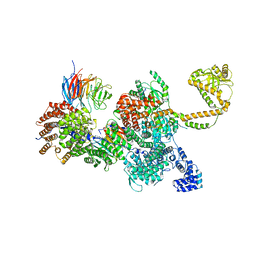 | | 80S-bound human SKI complex in the closed state | | Descriptor: | Helicase SKI2W, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3'), Tetratricopeptide repeat protein 37, ... | | Authors: | Koegel, A, Keidel, A, Bonneau, F, Schaefer, I.B, Conti, E. | | Deposit date: | 2021-12-01 | | Release date: | 2022-02-09 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The human SKI complex regulates channeling of ribosome-bound RNA to the exosome via an intrinsic gatekeeping mechanism.
Mol.Cell, 82, 2022
|
|
7QDY
 
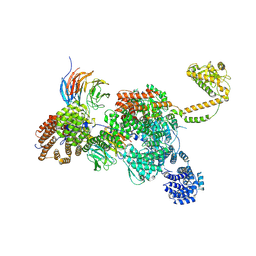 | | RNA-bound human SKI complex | | Descriptor: | Helicase SKI2W, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3'), Tetratricopeptide repeat protein 37, ... | | Authors: | Koegel, A, Keidel, A, Bonneau, F, Schaefer, I.B, Conti, E. | | Deposit date: | 2021-12-01 | | Release date: | 2022-02-09 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The human SKI complex regulates channeling of ribosome-bound RNA to the exosome via an intrinsic gatekeeping mechanism.
Mol.Cell, 82, 2022
|
|
4W8Z
 
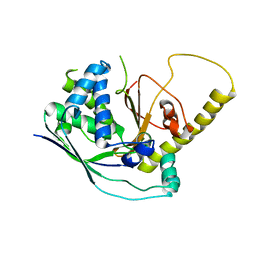 | |
4W8V
 
 | |
