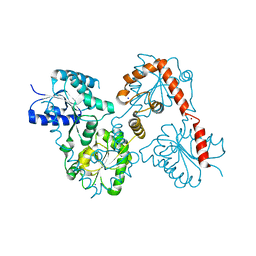
5W8N
 
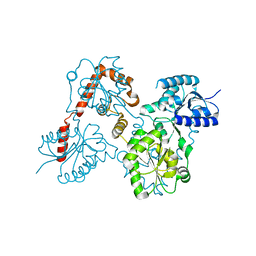 | |
5W8X
 
 | | Lipid A Disaccharide Synthase (LpxB)-7 solubilizing mutations-Bound to UDP | | Descriptor: | Lipid-A-disaccharide synthase, URIDINE-5'-DIPHOSPHATE | | Authors: | Bohl, T.E, Aihara, H, Shi, K, Lee, J.K. | | Deposit date: | 2017-06-22 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of lipid A disaccharide synthase LpxB from Escherichia coli.
Nat Commun, 9, 2018
|
|
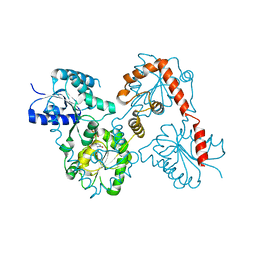
5WLY
 
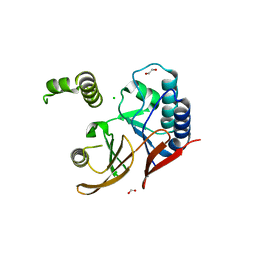 | | E. coli LpxH- 8 mutations | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Bohl, T.E, Aihara, H, Shi, K, Lee, J.K. | | Deposit date: | 2017-07-28 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The substrate-binding cap of the UDP-diacylglucosamine pyrophosphatase LpxH is highly flexible, enabling facile substrate binding and product release.
J. Biol. Chem., 293, 2018
|
|
5W8S
 
 | | Lipid A Disaccharide Synthase (LpxB)-7 solubilizing mutations | | Descriptor: | LITHIUM ION, Lipid-A-disaccharide synthase, SODIUM ION | | Authors: | Bohl, T.E, Aihara, H, Shi, K, Lee, J.K. | | Deposit date: | 2017-06-22 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of lipid A disaccharide synthase LpxB from Escherichia coli.
Nat Commun, 9, 2018
|
|
6E8C
 
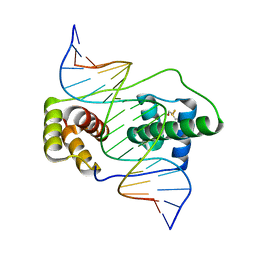 | | Crystal structure of the double homeodomain of DUX4 in complex with DNA | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*CP*GP*C)-3'), Double homeobox protein 4 | | Authors: | Lee, J.K, Bosnakovski, D, Toso, E.A, Dinh, T, Banerjee, S, Bohl, T.E, Shi, K, Kurahashi, K, Kyba, M, Aihara, H. | | Deposit date: | 2018-07-27 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal Structure of the Double Homeodomain of DUX4 in Complex with DNA.
Cell Rep, 25, 2018
|
|
6VOY
 
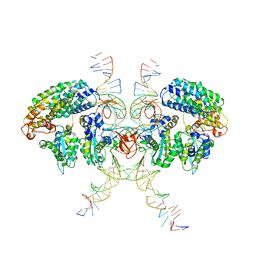 | | Cryo-EM structure of HTLV-1 instasome | | Descriptor: | DNA (25-MER), DNA (5'-D(P*AP*CP*AP*CP*AP*CP*TP*TP*GP*AP*CP*TP*AP*GP*GP*GP*TP*G)-3'), DNA-binding protein 7d, ... | | Authors: | Bhatt, V, Shi, K, Sundborger, A, Aihara, H. | | Deposit date: | 2020-02-01 | | Release date: | 2020-07-01 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of host protein hijacking in human T-cell leukemia virus integration.
Nat Commun, 11, 2020
|
|
6DRT
 
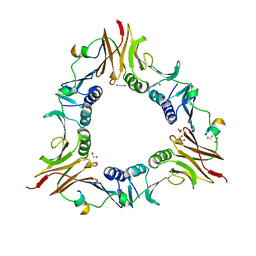 | |
6DT1
 
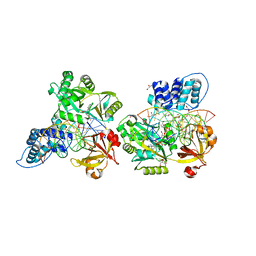 | | Crystal structure of the ligase from bacteriophage T4 complexed with DNA intermediate | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Shi, K, Aihara, H. | | Deposit date: | 2018-06-14 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | T4 DNA ligase structure reveals a prototypical ATP-dependent ligase with a unique mode of sliding clamp interaction.
Nucleic Acids Res., 46, 2018
|
|
5WFY
 
 | |
