2EU1
 
 | | Crystal structure of the chaperonin GroEL-E461K | | Descriptor: | GROEL | | Authors: | Cabo-Bilbao, A, Spinelli, S, Sot, B, Agirre, J, Mechaly, A.E, Muga, A, Guerin, D.M.A. | | Deposit date: | 2005-10-28 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Crystal structure of the temperature-sensitive and allosteric-defective chaperonin GroEL(E461K).
J.Struct.Biol., 155, 2006
|
|
3JBH
 
 | | TWO HEAVY MEROMYOSIN INTERACTING-HEADS MOTIFS FLEXIBLE DOCKED INTO TARANTULA THICK FILAMENT 3D-MAP ALLOWS IN DEPTH STUDY OF INTRA- AND INTERMOLECULAR INTERACTIONS | | Descriptor: | MYOSIN 2 ESSENTIAL LIGHT CHAIN STRIATED MUSCLE, MYOSIN 2 HEAVY CHAIN STRIATED MUSCLE, MYOSIN 2 REGULATORY LIGHT CHAIN STRIATED MUSCLE | | Authors: | Alamo, L, Qi, D, Wriggers, W, Pinto, A, Zhu, J, Bilbao, A, Gillilan, R.E, Hu, S, Padron, R. | | Deposit date: | 2015-09-01 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Conserved Intramolecular Interactions Maintain Myosin Interacting-Heads Motifs Explaining Tarantula Muscle Super-Relaxed State Structural Basis.
J. Mol. Biol., 428, 2016
|
|
2YEY
 
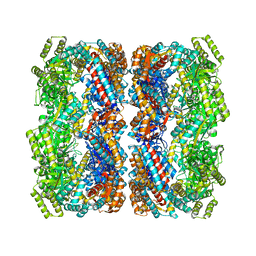 | | Crystal structure of the allosteric-defective chaperonin GroEL E434K mutant | | Descriptor: | 60 KDA CHAPERONIN | | Authors: | Cabo-Bilbao, A, Mechaly, A.E, Agirre, J, Spinelli, S, Sot, B, Muga, A, Guerin, D.M.A. | | Deposit date: | 2011-03-31 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Crystal Structure of the Temperature-Sensitive and Allosteric-Defective Chaperonin Groele461K.
J.Struct.Biol., 155, 2006
|
|
6YU6
 
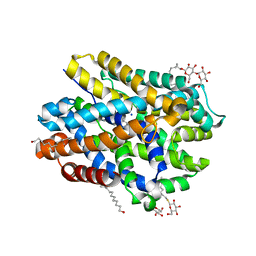 | | Crystal structure of MhsT in complex with L-leucine | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, LEUCINE, SODIUM ION, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
6YU4
 
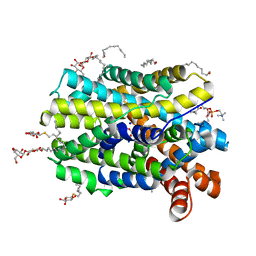 | | Crystal structure of MhsT in complex with L-4F-phenylalanine | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 4-FLUORO-L-PHENYLALANINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
6YU5
 
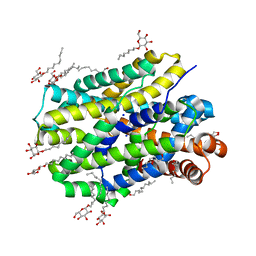 | | Crystal structure of MhsT in complex with L-valine | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, SODIUM ION, Sodium-dependent transporter, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
6YU7
 
 | | Crystal structure of MhsT in complex with L-tyrosine | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, SODIUM ION, Sodium-dependent transporter, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
6YU3
 
 | | Crystal structure of MhsT in complex with L-phenylalanine | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, GLYCEROL, PHENYLALANINE, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
6YU2
 
 | | Crystal structure of MhsT in complex with L-isoleucine | | Descriptor: | ISOLEUCINE, SODIUM ION, Sodium-dependent transporter, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
3RWV
 
 | | Crystal Structure of apo-form of Human Glycolipid Transfer Protein at 1.5 A resolution | | Descriptor: | Glycolipid transfer protein, SULFATE ION | | Authors: | Samygina, V, Cabo-Bilbao, A, Popov, A.N, Ochoa-Lizarralde, B, Goni-de-Cerio, F, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2011-05-09 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein.
Structure, 19, 2011
|
|
3S0K
 
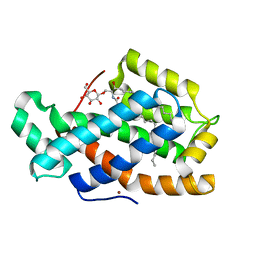 | | Crystal Structure of Human Glycolipid Transfer Protein complexed with glucosylceramide containing oleoyl acyl chain (18:1) | | Descriptor: | (9Z)-N-[(2S,3R,4E)-1-(beta-D-glucopyranosyloxy)-3-hydroxyoctadec-4-en-2-yl]octadec-9-enamide, Glycolipid transfer protein, NICKEL (II) ION | | Authors: | Cabo-Bilbao, A, Samygina, V, Popov, A.N, Ochoa-Lizarralde, B, Goni-de-Cerio, F, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2011-05-13 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein.
Structure, 19, 2011
|
|
3S0I
 
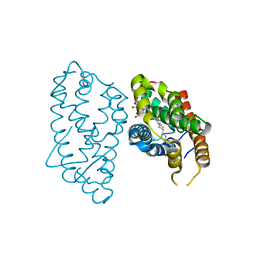 | | Crystal Structure of D48V mutant of Human Glycolipid Transfer Protein complexed with 3-O-sulfo galactosylceramide containing nervonoyl acyl chain | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, Glycolipid transfer protein | | Authors: | Samygina, V, Popov, A.N, Cabo-Bilbao, A, Ochoa-Lizarralde, B, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2011-05-13 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein.
Structure, 19, 2011
|
|
3RZN
 
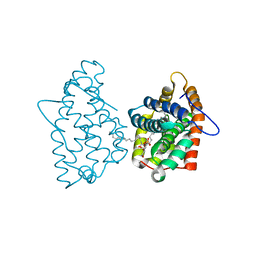 | | Crystal Structure of Human Glycolipid Transfer Protein complexed with 3-O-sulfo-galactosylceramide containing nervonoyl acyl chain (24:1) | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, Glycolipid transfer protein | | Authors: | Samygina, V, Cabo-Bilbao, A, Popov, A.N, Ochoa-Lizarralde, B, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2011-05-12 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein.
Structure, 19, 2011
|
|
4GHS
 
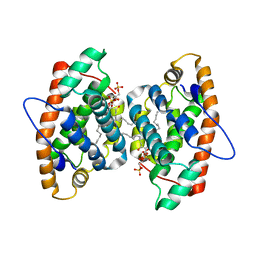 | | Crystal structure of human GLTP bound with 12:0 disulfatide (orthorombic form; two subunits in asymmetric unit) | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-1-[(3,6-di-O-sulfo-beta-D-galactopyranosyl)oxy]-3-hydroxyoctadec-4-en-2-yl}dodecanamide | | Authors: | Samygina, V.R, Popov, A.N, Goni-de-Cerio, F, Cabo-Bilbao, A, Malinina, L. | | Deposit date: | 2012-08-08 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GJQ
 
 | | Crystal structure of human GLTP bound with 12:0 monosulfatide (orthorhombic form;two subunits in asymmetric unit) | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, Glycolipid transfer protein, HEXANE | | Authors: | Cabo-Bilbao, A, Goni-de-Cerio, F, Samygina, V.R, Ochoa-Lizarralde, B, Popov, A.N, Malinina, L. | | Deposit date: | 2012-08-10 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GH0
 
 | | Crystal structure of D48V mutant of human GLTP bound with 12:0 monosulfatide | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-3-hydroxy-1-[(3-O-sulfo-beta-D-galactopyranosyl)oxy]octadec-4-en-2-yl}dodecanamide, PENTADECANE | | Authors: | Samygina, V.R, Cabo-Bilbao, A, Ochoa-Lizarralde, B, Popov, A.N, Malinina, L. | | Deposit date: | 2012-08-07 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GVT
 
 | | Crystal structure of D48V mutant of human GLTP bound with 12:0 disulfatide (hexagonal form) | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-1-[(3,6-di-O-sulfo-beta-D-galactopyranosyl)oxy]-3-hydroxyoctadec-4-en-2-yl}dodecanamide | | Authors: | Samygina, V.R, Cabo-Bilbao, A, Goni-de-Cerio, F, Popov, A.N, Malinina, L. | | Deposit date: | 2012-08-31 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GXG
 
 | | Crystal structure of human GLTP bound with 12:0 monosulfatide (orthorhombic form; four subunits in asymmetric unit) | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-3-hydroxy-1-[(3-O-sulfo-beta-D-galactopyranosyl)oxy]octadec-4-en-2-yl}dodecanamide | | Authors: | Samygina, V.R, Cabo-Bilbao, A, Goni-de-Cerio, F, Popov, A.N, Malinina, L. | | Deposit date: | 2012-09-04 | | Release date: | 2013-04-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5TBY
 
 | | HUMAN BETA CARDIAC HEAVY MEROMYOSIN INTERACTING-HEADS MOTIF OBTAINED BY HOMOLOGY MODELING (USING SWISS-MODEL) OF HUMAN SEQUENCE FROM APHONOPELMA HOMOLOGY MODEL (PDB-3JBH), RIGIDLY FITTED TO HUMAN BETA-CARDIAC NEGATIVELY STAINED THICK FILAMENT 3D-RECONSTRUCTION (EMD-2240) | | Descriptor: | Myosin light chain 3, Myosin regulatory light chain 2, ventricular/cardiac muscle isoform, ... | | Authors: | ALAMO, L, WARE, J.S, PINTO, A, GILLILAN, R.E, SEIDMAN, J.G, SEIDMAN, C.E, PADRON, R. | | Deposit date: | 2016-09-13 | | Release date: | 2017-06-07 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Effects of myosin variants on interacting-heads motif explain distinct hypertrophic and dilated cardiomyopathy phenotypes.
Elife, 6, 2017
|
|
3RIC
 
 | | Crystal Structure of D48V||A47D mutant of Human Glycolipid Transfer Protein complexed with 3-O-sulfo-galactosylceramide containing nervonoyl acyl chain (24:1) | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, Glycolipid transfer protein | | Authors: | Samygina, V, Ochoa-Lizarralde, B, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2011-04-13 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein.
Structure, 19, 2011
|
|
4GIX
 
 | | Crystal structure of human GLTP bound with 12:0 disulfatide | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-1-[(3,6-di-O-sulfo-beta-D-galactopyranosyl)oxy]-3-hydroxyoctadec-4-en-2-yl}dodecanamide | | Authors: | Samygina, V.R, Ochoa-Lizarralde, B, Malinina, L. | | Deposit date: | 2012-08-09 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GHP
 
 | | Crystal Structure of D48V||A47D mutant of Human GLTP bound with 12:0 monosulfatide | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-3-hydroxy-1-[(3-O-sulfo-beta-D-galactopyranosyl)oxy]octadec-4-en-2-yl}dodecanamide | | Authors: | Samygina, V.R, Ochoa-Lizarralde, B, Popov, A.N, Malinina, L. | | Deposit date: | 2012-08-08 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GXD
 
 | | Crystal structure of D48V mutant of human GLTP bound with 12:0 disulfatide | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-1-[(3,6-di-O-sulfo-beta-D-galactopyranosyl)oxy]-3-hydroxyoctadec-4-en-2-yl}dodecanamide | | Authors: | Samygina, V.R, Ochoa-Lizarralde, B, Popov, A.N, Malinina, L. | | Deposit date: | 2012-09-04 | | Release date: | 2013-04-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4H2Z
 
 | | Crystal structure of human GLTP bound with 12:0 monosulfatide | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-3-hydroxy-1-[(3-O-sulfo-beta-D-galactopyranosyl)oxy]octadec-4-en-2-yl}dodecanamide, PENTADECANE, ... | | Authors: | Samygina, V.R, Ochoa-Lizarralde, B, Malinina, L. | | Deposit date: | 2012-09-13 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
