7QSU
 
 | |
7QST
 
 | |
7QSS
 
 | |
6RIZ
 
 | | Crystal structure of gp41-1 intein (C1A, F65W, D107C) | | Descriptor: | gp41-1 intein | | Authors: | Beyer, H.M, Lountos, G.T, Mikula, M.K, Wlodawer, A, Iwai, H. | | Deposit date: | 2019-04-25 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Convergence of the Hedgehog/Intein Fold in Different Protein Splicing Mechanisms.
Int J Mol Sci, 21, 2020
|
|
6RIX
 
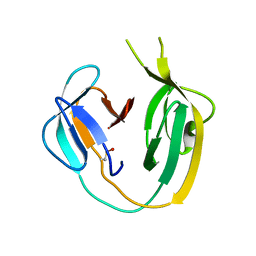 | | Crystal structure of MchDnaB-1 intein | | Descriptor: | CHLORIDE ION, Replicative DNA helicase | | Authors: | Beyer, H.M, Lountos, G.T, Mikula, M.K, Wlodawer, A, Iwai, H. | | Deposit date: | 2019-04-25 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.655 Å) | | Cite: | The Convergence of the Hedgehog/Intein Fold in Different Protein Splicing Mechanisms.
Int J Mol Sci, 21, 2020
|
|
6RIY
 
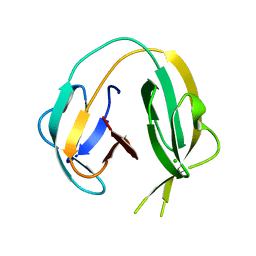 | | Crystal structure of MchDnaB-1 intein (N145AA) | | Descriptor: | CHLORIDE ION, Replicative DNA helicase | | Authors: | Beyer, H.M, Lountos, G.T, Mikula, M.K, Wlodawer, A, Iwai, H. | | Deposit date: | 2019-04-25 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | The Convergence of the Hedgehog/Intein Fold in Different Protein Splicing Mechanisms.
Int J Mol Sci, 21, 2020
|
|
6RPQ
 
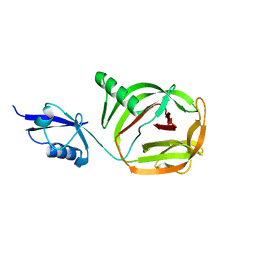 | | Crystal structure of PhoCDC21-1 intein | | Descriptor: | Ubiquitin-like protein SMT3,1108aa long hypothetical cell division control protein | | Authors: | Beyer, H.M, Mikula, K.M, Iwai, H. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.654 Å) | | Cite: | Crystal structures of CDC21-1 inteins from hyperthermophilic archaea reveal the selection mechanism for the highly conserved homing endonuclease insertion site.
Extremophiles, 23, 2019
|
|
6QAZ
 
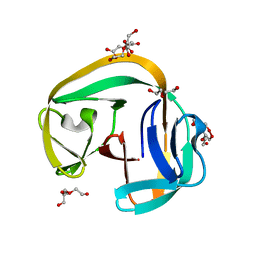 | | Crystal structure of gp41-1 intein | | Descriptor: | CITRATE ANION, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | Authors: | Mikula, K.M, Beyer, H.M, Li, M, Wlodawer, A, Iwai, H. | | Deposit date: | 2018-12-20 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | The crystal structure of the naturally split gp41-1 intein guides the engineering of orthogonal split inteins from cis-splicing inteins.
Febs J., 287, 2020
|
|
6RPP
 
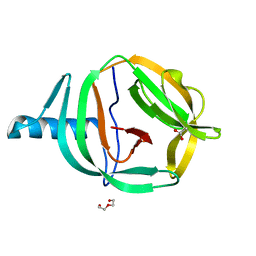 | | Crystal structure of PabCDC21-1 intein | | Descriptor: | ACETATE ION, Cell division control protein, DI(HYDROXYETHYL)ETHER | | Authors: | Mikula, K.M, Beyer, H.M, Iwai, H. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of CDC21-1 inteins from hyperthermophilic archaea reveal the selection mechanism for the highly conserved homing endonuclease insertion site.
Extremophiles, 23, 2019
|
|
8R2C
 
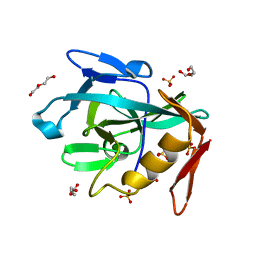 | | Crystal structure of the Vint domain from Tetrahymena thermophila | | Descriptor: | DI(HYDROXYETHYL)ETHER, SULFATE ION, von willebrand factor type A (VWA) domain was originally protein | | Authors: | Iwai, H, Beyer, H.M, Johannson, J.E, Li, M, Wlodawer, A. | | Deposit date: | 2023-11-03 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The three-dimensional structure of the Vint domain from Tetrahymena thermophila suggests a ligand-regulated cleavage mechanism by the HINT fold.
Febs Lett., 2024
|
|
7OEC
 
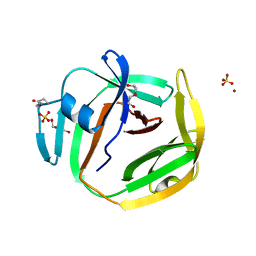 | | Crystal structure of an intein from a hyperthermophile | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA polymerase II large subunit, SULFATE ION, ... | | Authors: | Hannes, B, Hiltunen, M, Iwai, H. | | Deposit date: | 2021-05-03 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Mini-Intein Structures from Extremophiles Suggest a Strategy for Finding Novel Robust Inteins.
Microorganisms, 9, 2021
|
|
6SLY
 
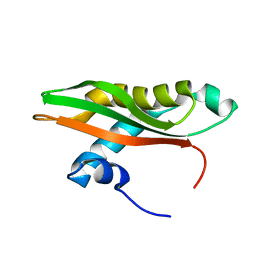 | |
