5NKL
 
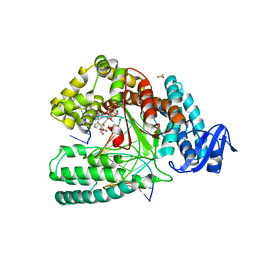 | | Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in a closed ternary complex with the artificial base pair dDs-dPxTP | | Descriptor: | ACETATE ION, DNA (5'-D(*AP*AP*AP*(DNU)P*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | Authors: | Betz, K, Marx, A, Diederichs, K, Hirao, I, Kimoto, M. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Expansion of the Genetic Alphabet with an Artificial Nucleobase Pair.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5O7T
 
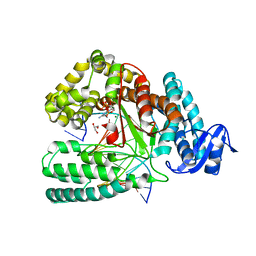 | | Crystal structure of KlenTaq mutant M747K in a closed ternary complex with a dG:dCTP base pair | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA polymerase I, thermostable, ... | | Authors: | Betz, K, Diederichs, K, Marx, A. | | Deposit date: | 2017-06-09 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the selective incorporation of an artificial nucleotide opposite a DNA adduct by a DNA polymerase.
Chem. Commun. (Camb.), 53, 2017
|
|
5OXJ
 
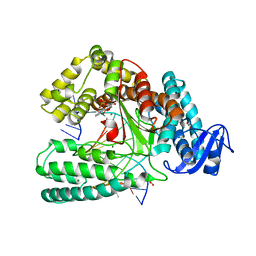 | | Crystal structure of KlenTaq mutant M747K in a closed ternary complex with a O6-MeG:BenziTP base pair | | Descriptor: | ACETATE ION, DNA polymerase I, thermostable, ... | | Authors: | Betz, K, Diederichs, K, Marx, A. | | Deposit date: | 2017-09-06 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the selective incorporation of an artificial nucleotide opposite a DNA adduct by a DNA polymerase.
Chem. Commun. (Camb.), 53, 2017
|
|
4C8M
 
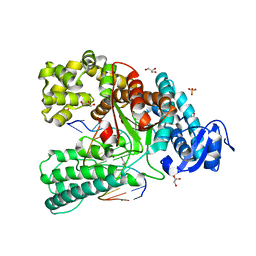 | | Binary complex of the large fragment of DNA polymerase I from Thermus Aquaticus with the aritificial base pair d5SICS-dNaM at the postinsertion site (sequence context 2) | | Descriptor: | GLYCEROL, LARGE FRAGMENT OF TAQ DNA POLYMERASE I, MAGNESIUM ION, ... | | Authors: | Betz, K, Malyshev, D.A, Lavergne, T, Welte, W, Diederichs, K, Romesberg, F.E, Marx, A. | | Deposit date: | 2013-10-01 | | Release date: | 2013-12-11 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (1.568 Å) | | Cite: | Structural Insights Into DNA Replication without Hydrogen Bonds.
J.Am.Chem.Soc., 135, 2013
|
|
4CCH
 
 | | Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in an open binary complex with d5SICS as templating nucleotide | | Descriptor: | 5'-D(*AP*AP*CP*LHOP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP* C)-3', 5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*DOC)-3', DNA POLYMERASE I, ... | | Authors: | Betz, K, Malyshev, D.A, Lavergne, T, Welte, W, Diederichs, K, Romesberg, F.E, Marx, A. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-11 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Insights Into DNA Replication without Hydrogen Bonds.
J.Am.Chem.Soc., 135, 2013
|
|
4C8N
 
 | | Binary complex of the large fragment of DNA polymerase I from Thermus Aquaticus with the aritificial base pair dNaM-d5SICS at the postinsertion site (sequence context 3) | | Descriptor: | LARGE FRAGMENT OF TAQ DNA POLYMERASE I, PRIMER, 5'-D(*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*LHOP)-3', ... | | Authors: | Betz, K, Malyshev, D.A, Lavergne, T, Welte, W, Diederichs, K, Romesberg, F.E, Marx, A. | | Deposit date: | 2013-10-01 | | Release date: | 2013-12-11 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural Insights Into DNA Replication without Hydrogen Bonds.
J.Am.Chem.Soc., 135, 2013
|
|
4C8O
 
 | | Binary complex of the large fragment of DNA polymerase I from Thermus Aquaticus with the aritificial base pair dNaM-d5SICS at the postinsertion site (sequence context 2) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*GP*CP*CP*AP*CP*GP*GP*CP*GP*CP*LHOP)-3', 5'-D(*TP*TP*CP*BMNP*GP*CP*GP*CP*CP*GP*TP*GP*GP*CP)-3', ... | | Authors: | Betz, K, Malyshev, D.A, Lavergne, T, Welte, W, Diederichs, K, Romesberg, F.E, Marx, A. | | Deposit date: | 2013-10-01 | | Release date: | 2013-12-11 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Insights Into DNA Replication without Hydrogen Bonds.
J.Am.Chem.Soc., 135, 2013
|
|
4C8K
 
 | | Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in a partially closed complex with the artificial base pair d5SICS-dNaMTP | | Descriptor: | ((2R,3S,5R)-3-hydroxy-5-(3-methoxynaphthalen-2-yl)methyl-tetrahydrogen-triphosphate, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*AP*AP*C*LHOP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3', ... | | Authors: | Betz, K, Malyshev, D.A, Lavergne, T, Welte, W, Diederichs, K, Romesberg, F.E, Marx, A. | | Deposit date: | 2013-10-01 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural Insights Into DNA Replication without Hydrogen Bonds.
J.Am.Chem.Soc., 135, 2013
|
|
4C8L
 
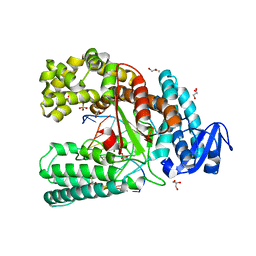 | | Binary complex of the large fragment of DNA polymerase I from Thermus Aquaticus with the artificial base pair dNaM-d5SICS at the postinsertion site (sequence context 1) | | Descriptor: | 5'-D(*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*LHOP)-3', 5'-D(*AP*GP*BMNP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP)-3', DNA POLYMERASE I, ... | | Authors: | Betz, K, Malyshev, D.A, Lavergne, T, Welte, W, Diederichs, K, Romesberg, F.E, Marx, A. | | Deposit date: | 2013-10-01 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights Into DNA Replication without Hydrogen Bonds.
J.Am.Chem.Soc., 135, 2013
|
|
3SV4
 
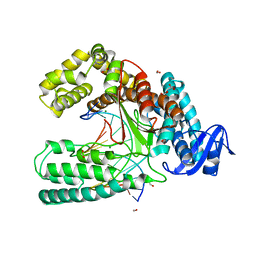 | | Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in an open binary complex with dT as templating nucleobase | | Descriptor: | (5'-D(*AP*AP*AP*TP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), DNA polymerase I, ... | | Authors: | Betz, K, Diederichs, K, Marx, A. | | Deposit date: | 2011-07-12 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | KlenTaq polymerase replicates unnatural base pairs by inducing a Watson-Crick geometry.
Nat.Chem.Biol., 8, 2012
|
|
3SYZ
 
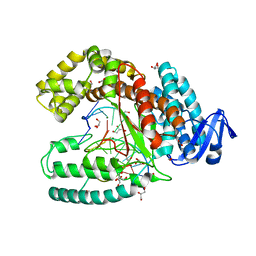 | | Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in an open binary complex with dNaM as templating nucleobase | | Descriptor: | (5'-D(*AP*AP*AP*(BMN)P*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), DNA polymerase I, ... | | Authors: | Betz, K, Diederichs, K, Marx, A. | | Deposit date: | 2011-07-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | KlenTaq polymerase replicates unnatural base pairs by inducing a Watson-Crick geometry.
Nat.Chem.Biol., 8, 2012
|
|
3SZ2
 
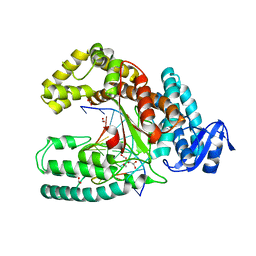 | | Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in an open binary complex with dG as templating nucleobase | | Descriptor: | (5'-D(*AP*AP*AP*GP*CP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DDG))-3'), DNA polymerase I, ... | | Authors: | Betz, K, Marx, A, Diederichs, K. | | Deposit date: | 2011-07-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | KlenTaq polymerase replicates unnatural base pairs by inducing a Watson-Crick geometry.
Nat.Chem.Biol., 8, 2012
|
|
3SV3
 
 | | Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in a closed ternary complex with the artificial base pair dNaM-d5SICSTP | | Descriptor: | (5'-D(*AP*AP*AP*(BMN)P*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), 2-{2-deoxy-5-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-erythro-pentofuranosyl}-6-methylisoquinoline-1(2H)-thione, ... | | Authors: | Betz, K, Diederichs, K, Marx, A. | | Deposit date: | 2011-07-12 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | KlenTaq polymerase replicates unnatural base pairs by inducing a Watson-Crick geometry.
Nat.Chem.Biol., 8, 2012
|
|
3M8R
 
 | | Crystal structure of the large fragment of DNA polymerase I from Thermus aquaticus in a closed ternary complex with trapped 4'-ethylated dTTP | | Descriptor: | 4'-ethylthymidine 5'-(tetrahydrogen triphosphate), ACETATE ION, DNA (5'-D(*AP*AP*AP*AP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Diederichs, K, Marx, A, Betz, K. | | Deposit date: | 2010-03-19 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of DNA polymerases caught processing size-augmented nucleotide probes.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
3M8S
 
 | | Crystal structure of the large fragment of DNA polymerase I from Thermus aquaticus in a closed ternary complex with trapped 4'-methylated dTTP | | Descriptor: | 4'-methylthymidine 5'-(tetrahydrogen triphosphate), ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Diederichs, K, Marx, A, Betz, K. | | Deposit date: | 2010-03-19 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of DNA polymerases caught processing size-augmented nucleotide probes.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
3RTV
 
 | | Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in a closed ternary complex with natural primer/template DNA | | Descriptor: | (5'-D(*AP*AP*AP*GP*CP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DDG))-3'), 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Marx, A, Diederichs, K, Betz, K. | | Deposit date: | 2011-05-04 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | KlenTaq polymerase replicates unnatural base pairs by inducing a Watson-Crick geometry.
Nat.Chem.Biol., 8, 2012
|
|
7OMG
 
 | | Crystal structure of KOD DNA Polymerase in a ternary complex with an Uracil containing template | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Betz, K, Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2021-05-22 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for The Recognition of Deaminated Nucleobases by An Archaeal DNA Polymerase.
Chembiochem, 22, 2021
|
|
7OM3
 
 | | Crystal structure of KOD DNA Polymerase in a binary complex with Hypoxanthine containing template | | Descriptor: | 1,2-ETHANEDIOL, 21nt Template, BROMIDE ION, ... | | Authors: | Betz, K, Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2021-05-21 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Basis for The Recognition of Deaminated Nucleobases by An Archaeal DNA Polymerase.
Chembiochem, 22, 2021
|
|
7OMB
 
 | | Crystal structure of KOD DNA Polymerase in a ternary complex with a p/t duplex containing an extended 5' single stranded template overhang | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA polymerase, ... | | Authors: | Betz, K, Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2021-05-21 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Basis for The Recognition of Deaminated Nucleobases by An Archaeal DNA Polymerase.
Chembiochem, 22, 2021
|
|
7OWF
 
 | | KlenTaq DNA polymerase in a ternary complex with primer/template and the fluorescent nucleotide analog BFdUTP | | Descriptor: | DNA polymerase I, thermostable, GLYCEROL, ... | | Authors: | Betz, K, Srivatsan, S.G. | | Deposit date: | 2021-06-18 | | Release date: | 2022-06-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Microenvironment-Sensitive Fluorescent Nucleotide Probes from Benzofuran, Benzothiophene, and Selenophene as Substrates for DNA Polymerases.
J.Am.Chem.Soc., 144, 2022
|
|
3QN1
 
 | | Crystal structure of the PYR1 Abscisic Acid receptor in complex with the HAB1 type 2C phosphatase catalytic domain | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYR1, MANGANESE (II) ION, ... | | Authors: | Betz, K, Dupeux, F, Santiago, J, Marquez, J.A. | | Deposit date: | 2011-02-07 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of Abscisic Acid Signaling in Vivo by an Engineered Receptor-Insensitive Protein Phosphatase Type 2C Allele.
Plant Physiol., 156, 2011
|
|
5OMQ
 
 | | Ternary complex of 9N DNA polymerase in the replicative state with three metal ions in the active site | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA Primer, DNA Template, ... | | Authors: | Betz, K, Marx, A, Diederichs, K. | | Deposit date: | 2017-08-01 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Crystal structures of ternary complexes of archaeal B-family DNA polymerases.
PLoS ONE, 12, 2017
|
|
5OMV
 
 | | Ternary complex of 9N DNA polymerase in the replicative state with two metal ions in the active site | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA polymerase, DNA primer, ... | | Authors: | Betz, K, Marx, A, Diederichs, K. | | Deposit date: | 2017-08-01 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Crystal structures of ternary complexes of archaeal B-family DNA polymerases.
PLoS ONE, 12, 2017
|
|
4K8X
 
 | | Binary complex of 9N DNA polymerase in the replicative state | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-hydroxypropyl)-2-{(1E,3E,5E)-5-[1-(3-hydroxypropyl)-3,3-dimethyl-1,3-dihydro-2H-indol-2-ylidene]penta-1,3-dien-1-y l}-3,3-dimethyl-3H-indolium, CHLORIDE ION, ... | | Authors: | Betz, K, Diederichs, K, Marx, A. | | Deposit date: | 2013-04-19 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structures of KOD and 9N DNA Polymerases Complexed with Primer Template Duplex
Chembiochem, 14, 2013
|
|
3ZVU
 
 | | Structure of the PYR1 His60Pro mutant in complex with the HAB1 phosphatase and Abscisic acid | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABSCISIC ACID RECEPTOR PYR1, MANGANESE (II) ION, ... | | Authors: | Betz, K, Dupeux, F, Santiago, J, Rodriguez, P.L, Marquez, J.A. | | Deposit date: | 2011-07-27 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Thermodynamic Switch Modulates Abscisic Acid Receptor Sensitivity.
Embo J., 30, 2011
|
|
