3D4F
 
 | | SHV-1 beta-lactamase complex with LN1-255 | | Descriptor: | (3R)-4-{[(3,4-dihydroxyphenyl)acetyl]oxy}-N-(2-formylindolizin-3-yl)-3-sulfino-D-valine, Beta-lactamase SHV-1, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE | | Authors: | Pattanaik, P, Bethel, C.R, Hujer, A.M, Bonomo, R.A, Buynak, J.D, van den Akker, F. | | Deposit date: | 2008-05-14 | | Release date: | 2009-02-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Strategic Design of an Effective {beta}-Lactamase Inhibitor: LN-1-255, A 6-ALKYLIDENE-2'-SUBSTITUTED PENICILLIN SULFONE
J.Biol.Chem., 284, 2009
|
|
1TDL
 
 | | Structure of Ser130Gly SHV-1 beta-lactamase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase SHV-1, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE | | Authors: | Sun, T, Bethel, C.R, Bonomo, R.A, Knox, J.R. | | Deposit date: | 2004-05-23 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibitor-resistant class A beta-lactamases: consequences of the Ser130-to-Gly mutation seen in Apo and tazobactam structures of the SHV-1 variant
Biochemistry, 43, 2004
|
|
1TDG
 
 | | Complex of S130G SHV-1 beta-lactamase with tazobactam | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase SHV-1, ... | | Authors: | Sun, T, Bethel, C.R, Bonomo, R.A, Knox, J.R. | | Deposit date: | 2004-05-21 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibitor-resistant class A beta-lactamases: consequences of the Ser130-to-Gly mutation seen in Apo and tazobactam structures of the SHV-1 variant
Biochemistry, 43, 2004
|
|
3FV7
 
 | | OXA-24 beta-lactamase complex with SA4-44 inhibitor | | Descriptor: | (2S)-2-[[2-methanoyl-7-(methoxycarbonylamino)indolizin-3-yl]amino]-3-methyl-3-sulfino-butanoic acid, Beta-lactamase OXA-24 | | Authors: | Bou, G, Santillana, E, Sheri, A, Beceiro, A, Sampson, J.M, Kalp, M, Bethel, C.R, Distler, A.M, Drawz, S.M, Pagadala, S.R, Van den Akker, F, Bonomo, R.A, Romero, A, Buynak, J.D. | | Deposit date: | 2009-01-15 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, synthesis, and crystal structures of 6-alkylidene-2'-substituted penicillanic acid sulfones as potent inhibitors of Acinetobacter baumannii OXA-24 carbapenemase.
J.Am.Chem.Soc., 132, 2010
|
|
2G2W
 
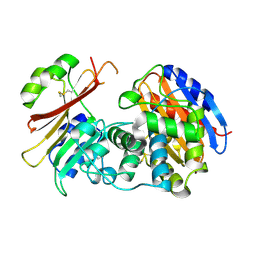 | | Crystal Structure of the SHV D104K Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) complex | | Descriptor: | Beta-lactamase SHV-1, Beta-lactamase inhibitory protein | | Authors: | Reynolds, K.A, Thomson, J.M, Corbett, K.D, Bethel, C.R, Berger, J.M, Kirsch, J.F, Bonomo, R.A, Handel, T.M. | | Deposit date: | 2006-02-16 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Computational Characterization of the SHV-1 beta-Lactamase-beta-Lactamase Inhibitor Protein Interface.
J.Biol.Chem., 281, 2006
|
|
2G2U
 
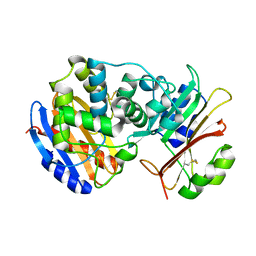 | | Crystal Structure of the SHV-1 Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) complex | | Descriptor: | Beta-lactamase SHV-1, Beta-lactamase inhibitory protein | | Authors: | Reynolds, K.A, Thomson, J.M, Corbett, K.D, Bethel, C.R, Berger, J.M, Kirsch, J.F, Bonomo, R.A, Handel, T.M. | | Deposit date: | 2006-02-16 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Computational Characterization of the SHV-1 beta-Lactamase-beta-Lactamase Inhibitor Protein Interface.
J.Biol.Chem., 281, 2006
|
|
2OV5
 
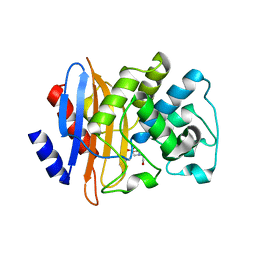 | | Crystal structure of the KPC-2 carbapenemase | | Descriptor: | BICINE, Carbapenemase | | Authors: | Ke, W, Bethel, C.R, Thomson, J.M, Bonomo, R.A, van den Akker, F. | | Deposit date: | 2007-02-13 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of KPC-2: Insights into Carbapenemase Activity in Class A beta-Lactamases.
Biochemistry, 46, 2007
|
|
4R3B
 
 | |
7BDS
 
 | |
7BDR
 
 | | Structure of CTX-M-15 E166Q mutant crystallised in the presence of tazobactam (AAI101) | | Descriptor: | Beta-lactamase, CHLORIDE ION, SODIUM ION, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-12-22 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | Penicillanic Acid Sulfones Inactivate the Extended-Spectrum beta-Lactamase CTX-M-15 through Formation of a Serine-Lysine Cross-Link: an Alternative Mechanism of beta-Lactamase Inhibition.
Mbio, 13, 2022
|
|
6MPQ
 
 | | 1.95 Ang crystal structure of OXA-24/40 beta-lactamase in complex the inhibitor ETX2514 | | Descriptor: | (2S,5R)-1-formyl-3-methyl-5-[(sulfooxy)amino]-1,2,5,6-tetrahydropyridine-2-carboxamide, Beta-lactamase, CHLORIDE ION | | Authors: | van den Akker, F, Kumar, V. | | Deposit date: | 2018-10-08 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Targeting Multidrug-ResistantAcinetobacterspp.: Sulbactam and the Diazabicyclooctenone beta-Lactamase Inhibitor ETX2514 as a Novel Therapeutic Agent.
MBio, 10, 2019
|
|
4X6T
 
 | | M.tuberculosis betalactamase complexed with inhibitor EC19 | | Descriptor: | 3-[(2R)-2-(dihydroxyboranyl)-2-{[(2R)-2-{[(4-ethyl-2,3-dioxo-3,4-dihydropyrazin-1(2H)-yl)carbonyl]amino}-2-(4-hydroxyphenyl)acetyl]amino}ethyl]benzoic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Hazra, S. | | Deposit date: | 2014-12-09 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inhibiting the beta-Lactamase of Mycobacterium tuberculosis (Mtb) with Novel Boronic Acid Transition-State Inhibitors (BATSIs).
ACS Infect Dis, 1, 2015
|
|
2ZD8
 
 | | SHV-1 class A beta-lactamase complexed with meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase SHV-1, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE | | Authors: | Nukaga, M, Knox, J.R, Hujer, A, Bonomo, R.A. | | Deposit date: | 2007-11-20 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Inhibition of class A beta-lactamases by carbapenems: crystallographic observation of two conformations of meropenem in SHV-1.
J.Am.Chem.Soc., 130, 2008
|
|
3RXW
 
 | | KPC-2 carbapenemase in complex with PSR3-226 | | Descriptor: | (2S,3R)-4-(2-amino-2-oxoethoxy)-3-(dihydroxy-lambda~4~-sulfanyl)-3-methyl-4-oxo-2-{[(1E)-3-oxoprop-1-en-1-yl]amino}butanoic acid, CITRIC ACID, Carbepenem-hydrolyzing beta-lactamase KPC | | Authors: | Ke, W, van den Akker, F. | | Deposit date: | 2011-05-10 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal structures of KPC-2 {beta}-lactamase in complex with 3-nitrophenyl boronic acid and the penam sulfone PSR-3-226.
Antimicrob.Agents Chemother., 56, 2012
|
|
3RXX
 
 | | KPC-2 carbapenemase in complex with 3-NPBA | | Descriptor: | 3-NITROPHENYLBORONIC ACID, Carbepenem-hydrolyzing beta-lactamase KPC | | Authors: | Ke, W, van den Akker, F. | | Deposit date: | 2011-05-10 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structures of KPC-2 {beta}-lactamase in complex with 3-nitrophenyl boronic acid and the penam sulfone PSR-3-226.
Antimicrob.Agents Chemother., 56, 2012
|
|
6OP6
 
 | | Structure of VIM-20 in the reduced state | | Descriptor: | Metallo-beta-lactamase VIM-20, SODIUM ION, ZINC ION | | Authors: | Page, R.C, Shurina, B.A, Montgomery, J.S, Orischak, M.G, Nix, J.C. | | Deposit date: | 2019-04-24 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A Single Salt Bridge in VIM-20 Increases Protein Stability and Antibiotic Resistance under Low-Zinc Conditions.
Mbio, 10, 2019
|
|
6OP7
 
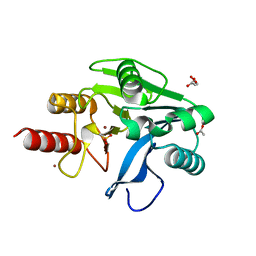 | | Structure of oxidized VIM-20 | | Descriptor: | ACETATE ION, Metallo-beta-lactamase VIM-20, ZINC ION | | Authors: | Page, R.C, Shurina, B.A, Montgomery, J.S, Orischak, M.G, Nix, J.C. | | Deposit date: | 2019-04-24 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | A Single Salt Bridge in VIM-20 Increases Protein Stability and Antibiotic Resistance under Low-Zinc Conditions.
Mbio, 10, 2019
|
|
5WIH
 
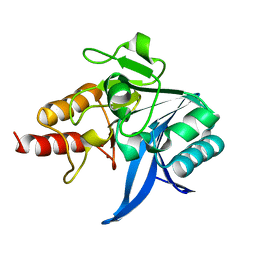 | |
5WIG
 
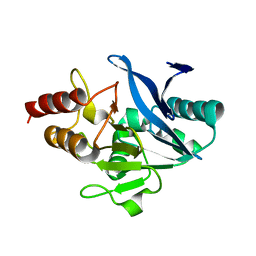 | |
7TB7
 
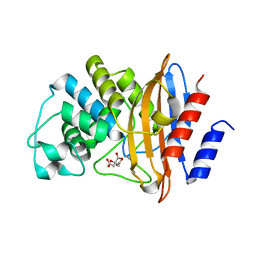 | | Crystal structure of D179N KPC-2 beta-lactamase | | Descriptor: | CITRIC ACID, Carbapenem-hydrolyzing beta-lactamase KPC | | Authors: | van den Akker, F, Alsenani, T. | | Deposit date: | 2021-12-21 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Structural Characterization of the D179N and D179Y Variants of KPC-2 beta-Lactamase: Omega-Loop Destabilization as a Mechanism of Resistance to Ceftazidime-Avibactam.
Antimicrob.Agents Chemother., 66, 2022
|
|
7TBX
 
 | | Crystal structure of D179Y KPC-2 beta-lactamase | | Descriptor: | Carbapenem-hydrolyzing beta-lactamase KPC | | Authors: | van den Akker, F, Alsenani, T. | | Deposit date: | 2021-12-22 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Structural Characterization of the D179N and D179Y Variants of KPC-2 beta-Lactamase: Omega-Loop Destabilization as a Mechanism of Resistance to Ceftazidime-Avibactam.
Antimicrob.Agents Chemother., 66, 2022
|
|
7TC1
 
 | |
7UTB
 
 | | KPC-2 CARBAPENEMASE IN COMPLEX WITH THE BORONIC ACID INHIBITOR MB_076 | | Descriptor: | Carbapenem-hydrolyzing beta-lactamase KPC, [(1~{R})-1-[2-[(5-azanyl-1,3,4-thiadiazol-2-yl)sulfanyl]ethanoylamino]-2-(4-carboxy-1,2,3-triazol-1-yl)ethyl]-$l^{3}-oxidanyl-bis(oxidanyl)boron | | Authors: | van den Akker, F, Alsenani, T.A. | | Deposit date: | 2022-04-26 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Boronic Acid Transition State Inhibitors as Potent Inactivators of KPC and CTX-M beta-Lactamases: Biochemical and Structural Analyses.
Antimicrob.Agents Chemother., 67, 2023
|
|
6VOT
 
 | | Crystal structure of Pseudomonas aerugonisa PBP3 complexed to gamma-lactam YU253434 | | Descriptor: | 1-[(2S)-2-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(2-carboxypropan-2-yl)oxy]imino}acetyl]amino}-3-oxopropyl]-4-{[2-(5,6 -dihydroxy-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl]carbamoyl}-2,5-dihydro-1H-pyrazole-3-carboxylic acid, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | van den Akker, F. | | Deposit date: | 2020-01-31 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A gamma-Lactam Siderophore Antibiotic Effective against Multidrug-Resistant Gram-Negative Bacilli.
J.Med.Chem., 63, 2020
|
|
6VJE
 
 | | Crystal structure of Pseudomonas aeruginosa penicillin-binding protein 3 (PBP3) complexed with ceftobiprole | | Descriptor: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyr rolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, CHLORIDE ION, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | van den Akker, F, Kumar, V. | | Deposit date: | 2020-01-15 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Insights into Ceftobiprole Inhibition of Pseudomonas aeruginosa Penicillin-Binding Protein 3.
Antimicrob.Agents Chemother., 64, 2020
|
|
