5G6U
 
 | | Crystal structure of langerin carbohydrate recognition domain with GlcNS6S | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Porkolab, V, Chabrol, E, Varga, N, Ordanini, S, Sutkeviciute, I, Thepaut, M, Bernardi, A, Fieschi, F. | | Deposit date: | 2016-07-21 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | Rational-Differential Design of Highly Specific Glycomimetic Ligands: Targeting DC-SIGN and Excluding Langerin Recognition.
ACS Chem. Biol., 13, 2018
|
|
2XR6
 
 | | Crystal structure of the complex of the carbohydrate recognition domain of human DC-SIGN with pseudo trimannoside mimic. | | Descriptor: | 2-AZIDOETHANOL, CALCIUM ION, CD209 ANTIGEN, ... | | Authors: | Thepaut, M, Suitkeviciute, I, Sattin, S, Reina, J, Bernardi, A, Fieschi, F. | | Deposit date: | 2010-09-10 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Unique Dc-Sign Clustering Activity of a Small Glycomimetic: A Lesson for Ligand Design.
Acs Chem.Biol., 9, 2014
|
|
4WX4
 
 | | Crystal structure of adenovirus 8 protease in complex with a nitrile inhibitor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCINE, N-[(2-cyanopyrimidin-4-yl)methyl]-3-[2-(3,5-dichlorophenyl)-2-methylpropanoyl]-4-methoxybenzamide, ... | | Authors: | Grosche, P, Sirockin, F, Mac Sweeney, A, Ramage, P, Erbel, P, Melkko, S, Bernardi, A, Hughes, N, Ellis, D, Combrink, K, Jarousse, N, Altmann, E. | | Deposit date: | 2014-11-13 | | Release date: | 2015-01-14 | | Last modified: | 2015-01-28 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Structure-based design and optimization of potent inhibitors of the adenoviral protease.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4WX6
 
 | | Crystal structure of human adenovirus 8 protease with an irreversible vinyl sulfone inhibitor | | Descriptor: | N-[(2S)-2-(3,5-dichlorophenyl)-2-(ethylamino)acetyl]-3-methyl-L-valyl-N-[3-(methylsulfonyl)propyl]glycinamide, PVI, Protease | | Authors: | Grosche, P, Sirockin, F, Mac Sweeney, A, Ramage, P, Erbel, P, Melkko, S, Bernardi, A, Hughes, N, Ellis, D, Combrink, K, Jarousse, N, Altmann, E. | | Deposit date: | 2014-11-13 | | Release date: | 2015-01-14 | | Last modified: | 2015-01-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-based design and optimization of potent inhibitors of the adenoviral protease.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4WX7
 
 | | Crystal structure of adenovirus 8 protease with a nitrile inhibitor | | Descriptor: | 3-[2-(3,5-dichlorophenyl)-2-methylpropanoyl]-N-(2-{[(2Z)-2-iminoethyl]amino}-2-oxoethyl)-4-methoxybenzamide, PVI, Protease | | Authors: | Grosche, P, Sirockin, F, Mac Sweeney, A, Ramage, P, Erbel, P, Melkko, S, Bernardi, A, Hughes, N, Ellis, D, Combrink, K, Jarousse, N, Altmann, E. | | Deposit date: | 2014-11-13 | | Release date: | 2015-01-14 | | Last modified: | 2015-01-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design and optimization of potent inhibitors of the adenoviral protease.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
6GHV
 
 | | Structure of a DC-SIGN CRD in complex with high affinity glycomimetic. | | Descriptor: | CALCIUM ION, CD209 antigen, CHLORIDE ION, ... | | Authors: | Thepaut, M, Achilli, S, Medve, L, Bernardi, A, Fieschi, F. | | Deposit date: | 2018-05-09 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enhancing Potency and Selectivity of a DC-SIGN Glycomimetic Ligand by Fragment-Based Design: Structural Basis.
Chemistry, 25, 2019
|
|
2XR5
 
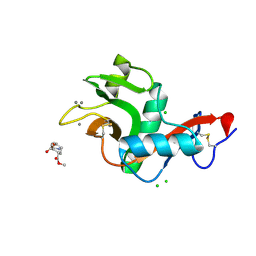 | | Crystal structure of the complex of the carbohydrate recognition domain of human DC-SIGN with pseudo dimannoside mimic. | | Descriptor: | CALCIUM ION, CD209 ANTIGEN, CHLORIDE ION, ... | | Authors: | Thepaut, M, Suitkeviciute, I, Sattin, S, Reina, J, Bernardi, A, Fieschi, F. | | Deposit date: | 2010-09-10 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure of a Glycomimetic Ligand in the Carbohydrate Recognition Domain of C-Type Lectin Dc-Sign. Structural Requirements for Selectivity and Ligand Design.
J.Am.Chem.Soc., 135, 2013
|
|
4PIS
 
 | | Crystal structure of human adenovirus 8 protease in complex with a nitrile inhibitor | | Descriptor: | N~2~-[(2R)-2-(3,5-dichlorophenyl)-2-(dimethylamino)acetyl]-N-({2-[(Z)-iminomethyl]pyrimidin-4-yl}methyl)-L-isoleucinamide, PVI, Protease | | Authors: | Mac Sweeney, A, Grosche, P, Ellis, D, Combrink, K, Erbel, P, Hughes, N, Sirockin, F, Melkko, S, Bernardi, A, Ramage, P, Jarousse, N, Altmann, E. | | Deposit date: | 2014-05-09 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and structure-based optimization of adenain inhibitors.
Acs Med.Chem.Lett., 5, 2014
|
|
4PIE
 
 | | Crystal structure of human adenovirus 2 protease a substrate based nitrile inhibitor | | Descriptor: | ACETATE ION, N-{(2S)-2-(3-chlorophenyl)-2-[(methylsulfonyl)amino]acetyl}-L-phenylalanyl-N-[(2Z)-2-iminoethyl]glycinamide, Pre-protein VI, ... | | Authors: | Mac Sweeney, A, Grosche, P, Ellis, D, Combrink, C, Erbel, C, Hughes, N, Sirockin, F, Melkko, S, Bernardi, A, Ramage, P, Jarousse, N, Altmann, E. | | Deposit date: | 2014-05-08 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery and structure-based optimization of adenain inhibitors.
Acs Med.Chem.Lett., 5, 2014
|
|
4PID
 
 | | Crystal structure of human adenovirus 2 protease with a weak pyrimidine nitrile inhibitor | | Descriptor: | ACETATE ION, N-benzyl-2-[(Z)-iminomethyl]pyrimidine-5-carboxamide, Pre-protein VI, ... | | Authors: | Mac Sweeney, A, Grosche, P, Ellis, D, Combrink, K, Erbel, P, Hughes, N, Sirockin, F, Melkko, S, Bernardi, A, Ramage, P, Jarousse, N, Altmann, E. | | Deposit date: | 2014-05-08 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Discovery and structure-based optimization of adenain inhibitors.
Acs Med.Chem.Lett., 5, 2014
|
|
4PIQ
 
 | | Crystal structure of human adenovirus 8 protease with a nitrile inhibitor | | Descriptor: | N-[(3,5-dichlorophenyl)acetyl]-L-threonyl-N-[(2Z)-2-iminoethyl]glycinamide, PVI, Protease | | Authors: | Mac Sweeney, A, Grosche, P, Ellis, D, Combrink, K, Erbel, P, Hughes, N, Sirockin, F, Melkko, S, Bernardi, A, Ramage, P, Jarousse, N, Altmann, E. | | Deposit date: | 2014-05-09 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery and structure-based optimization of adenain inhibitors.
Acs Med.Chem.Lett., 5, 2014
|
|
7BFY
 
 | |
6ZZW
 
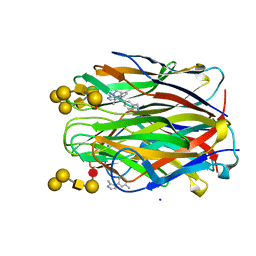 | | Structure of the N terminal domain of Bc2L-C lectin (1-131) in complex with Globo H (H-type 3) and CAS No 912569-62-1 | | Descriptor: | Lectin, SODIUM ION, [3-(2-methylimidazol-1-yl)phenyl]methanamine, ... | | Authors: | Lal, K, Bermeo, R, Imberty, A, Varrot, A. | | Deposit date: | 2020-08-05 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Prediction and Validation of a Druggable Site on Virulence Factor of Drug Resistant Burkholderia cenocepacia*.
Chemistry, 27, 2021
|
|
8BRO
 
 | | Structure of the N-terminal domain of BC2L-C lectin (1-131) in complex with a synthetic beta-fucosylamide | | Descriptor: | 5-[3-(aminomethyl)phenyl]-~{N}-[(2~{S},3~{S},4~{R},5~{S},6~{S})-6-methyl-3,4,5-tris(oxidanyl)oxan-2-yl]furan-2-carboxamide, Lectin | | Authors: | Varrot, A, Lunstrom, J. | | Deposit date: | 2022-11-23 | | Release date: | 2023-02-08 | | Last modified: | 2023-02-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification of New L-Fucosyl and L-Galactosyl Amides as Glycomimetic Ligands of TNF Lectin Domain of BC2L-C from Burkholderia cenocepacia.
Molecules, 28, 2023
|
|
7OLU
 
 | |
7OLW
 
 | |
6TIG
 
 | | Structure of the N terminal domain of Bc2L-C lectin (1-131) in complex with Globo H (H-type 3) antigen | | Descriptor: | Lectin, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-alpha-D-galactopyranose, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose | | Authors: | Varrot, A, Bermeo, R. | | Deposit date: | 2019-11-22 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | BC2L-C N-Terminal Lectin Domain Complexed with Histo Blood Group Oligosaccharides Provides New Structural Information.
Molecules, 25, 2020
|
|
6TID
 
 | |
3EDG
 
 | |
3EDI
 
 | |
3EDH
 
 | |
3V4O
 
 | |
3V4L
 
 | |
3V55
 
 | | Human MALT1 (334-719) in its ligand free form | | Descriptor: | Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | Renatus, M, Wiesmann, C. | | Deposit date: | 2011-12-16 | | Release date: | 2012-03-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural Determinants of MALT1 Protease Activity.
J.Mol.Biol., 419, 2012
|
|
5LZI
 
 | | Porcine heat-labile enterotoxin R13H in complex with inhibitor MM146 | | Descriptor: | (2~{R},4~{S},5~{R},6~{R})-5-acetamido-2-[4-[(2~{R})-3-[2-[(2~{S},3~{R},4~{R},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]ethylamino]-3-oxidanylidene-2-(2-phenylethanoylamino)propyl]-1,2,3-triazol-1-yl]-4-oxidanyl-6-[(1~{R},2~{R})-1,2,3-tris(oxidanyl)propyl]oxane-2-carboxylic acid, (2~{R},4~{S},5~{R},6~{R})-5-acetamido-2-[4-[(2~{S})-3-[2-[(2~{S},3~{R},4~{R},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]ethylamino]-3-oxidanylidene-2-(2-phenylethanoylamino)propyl]-1,2,3-triazol-1-yl]-4-oxidanyl-6-[(1~{R},2~{R})-1,2,3-tris(oxidanyl)propyl]oxane-2-carboxylic acid, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Heggelund, J.E, Mackenzie, A, Krengel, U. | | Deposit date: | 2016-09-29 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Towards new cholera prophylactics and treatment: Crystal structures of bacterial enterotoxins in complex with GM1 mimics.
Sci Rep, 7, 2017
|
|
