5N49
 
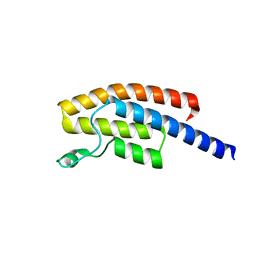 | | BRPF2 in complex with Compound 7 | | Descriptor: | 2-(1,3,6-trimethyl-2-oxidanylidene-benzimidazol-5-yl)benzo[de]isoquinoline-1,3-dione, Bromodomain-containing protein 1 | | Authors: | Bouche, L, Christ, C.D, Siegel, S, Fernandez-Montalvan, A.E, Holton, S.J, Fedorov, O, ter Laak, A, Sugawara, T, Stoeckigt, D, Tallant, C, Bennett, J, Monteiro, O, Saez, L.D, Siejka, P, Meier, J, Puetter, V, Weiske, J, Mueller, S, Huber, K.V.M, Hartung, I.V, Haendler, B. | | Deposit date: | 2017-02-10 | | Release date: | 2017-05-03 | | Last modified: | 2017-05-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Benzoisoquinolinediones as Potent and Selective Inhibitors of BRPF2 and TAF1/TAF1L Bromodomains.
J. Med. Chem., 60, 2017
|
|
7PSQ
 
 | | Crystal structure of S100A4 labeled with NU074381b. | | Descriptor: | (2~{R},4~{R})-1-ethanoyl-~{N}-naphthalen-1-yl-4-phenyl-pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Giroud, C, Szommer, T, Coxon, C, Monteiro, O, Christott, T, Bennett, J, Aitmakhanova, K, Raux, B, Newman, J, Elkins, J, Krojer, T, Arruda Bezerra, G, Koekemoer, L, Bountra, C, Von Delft, F, Brennan, P, Fedorov, O. | | Deposit date: | 2021-09-23 | | Release date: | 2022-10-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of S100A4 labeled with NU074381b.
To Be Published
|
|
5MG2
 
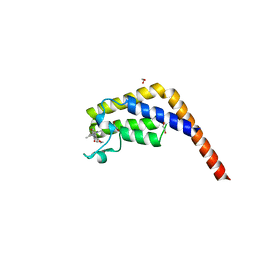 | | Crystal structure of the second bromodomain of human TAF1 in complex with BAY-299 chemical probe | | Descriptor: | 1,2-ETHANEDIOL, 6-(3-oxidanylpropyl)-2-(1,3,6-trimethyl-2-oxidanylidene-benzimidazol-5-yl)benzo[de]isoquinoline-1,3-dione, Transcription initiation factor TFIID subunit 1 | | Authors: | Tallant, C, Bouche, L, Holton, S.J, Fedorov, O, Siejka, P, Picaud, S, Krojer, T, Srikannathasan, V, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hartung, I.V, Haendler, B, Muller, S, Huber, K.V.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-11-20 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Benzoisoquinolinediones as Potent and Selective Inhibitors of BRPF2 and TAF1/TAF1L Bromodomains.
J. Med. Chem., 60, 2017
|
|
6I8L
 
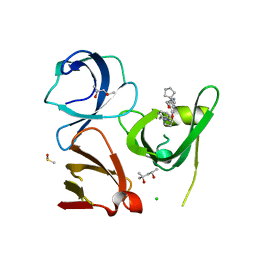 | | Crystal structure of Spindlin1 in complex with the inhibitor TD001851a | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 5'-(cyclopropylmethoxy)-6'-[3-(1,3-dihydroisoindol-2-yl)propoxy]spiro[cyclopentane-1,3'-indole]-2'-amine, ... | | Authors: | Johansson, C, Fagan, V, Brennan, P.E, Sorrell, F.J, Krojer, T, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2018-11-20 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A Chemical Probe for Tudor Domain Protein Spindlin1 to Investigate Chromatin Function.
J.Med.Chem., 62, 2019
|
|
6I8B
 
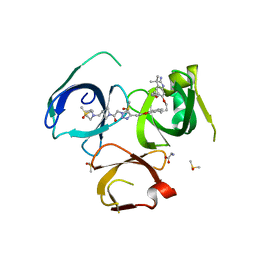 | | Crystal structure of Spindlin1 in complex with the inhibitor VinSpinIn | | Descriptor: | 2-[4-[2-[[2-[3-[2-azanyl-5-(cyclopropylmethoxy)-3,3-dimethyl-indol-6-yl]oxypropyl]-1,3-dihydroisoindol-5-yl]oxy]ethyl]-1,2,3-triazol-1-yl]-1-[4-(2-pyrrolidin-1-ylethyl)piperidin-1-yl]ethanone, DIMETHYL SULFOXIDE, GLYCINE, ... | | Authors: | Johansson, C, Fagan, V, Brennan, P.E, Sorrell, F.J, Krojer, T, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2018-11-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A Chemical Probe for Tudor Domain Protein Spindlin1 to Investigate Chromatin Function.
J.Med.Chem., 62, 2019
|
|
6HT1
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with SGC-iMLLT (compound 92) | | Descriptor: | 1,2-ETHANEDIOL, 1-methyl-~{N}-[2-[[(2~{S})-2-methylpyrrolidin-1-yl]methyl]-3~{H}-benzimidazol-5-yl]indazole-5-carboxamide, Protein ENL, ... | | Authors: | Heidenreich, D, Chaikuad, A, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-10-02 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of an MLLT1/3 YEATS Domain Chemical Probe.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6HT0
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with compound 94 | | Descriptor: | 1,2-ETHANEDIOL, 1-cyclopropyl-~{N}-[2-[[(2~{S})-2-methylpyrrolidin-1-yl]methyl]-3~{H}-benzimidazol-5-yl]indazole-5-carboxamide, Protein ENL, ... | | Authors: | Heidenreich, D, Chaikuad, A, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-10-02 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of an MLLT1/3 YEATS Domain Chemical Probe.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6I8Y
 
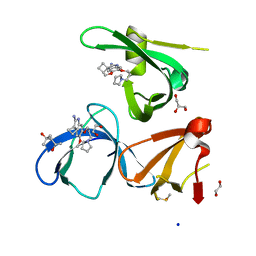 | | Crystal structure of Spindlin1 in complex with the Methyltransferase inhibitor A366 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 5'-methoxy-6'-[3-(pyrrolidin-1-yl)propoxy]spiro[cyclobutane-1,3'-indol]-2'-amine, ... | | Authors: | Srikannathasan, V, Johansson, C, Gileadi, C, Shrestha, L, Sorrell, F.J, Krojer, T, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2018-11-21 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A Chemical Probe for Tudor Domain Protein Spindlin1 to Investigate Chromatin Function.
J.Med.Chem., 62, 2019
|
|
7OL1
 
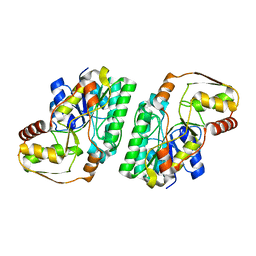 | |
8DKB
 
 | |
8PJ7
 
 | | MLLT3 in complex with compound PFI-6 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, PHOSPHATE ION, ... | | Authors: | Raux, B, Diaz-Saez, L, Huber, K.V.M, Fedorov, O, Owen, D.R, Londregan, A.T, Bountra, C, Edwards, A, Arrowsmith, C. | | Deposit date: | 2023-06-22 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Discovery of PFI-6, a small-molecule chemical probe for the YEATS domain of MLLT1 and MLLT3.
Bioorg.Med.Chem.Lett., 98, 2023
|
|
8PJI
 
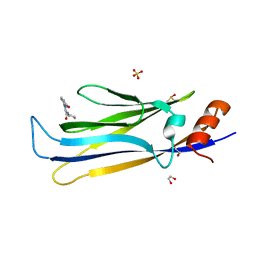 | | MLLT1 in complex with compound 10a | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Protein ENL, ... | | Authors: | Raux, B, Diaz-Saez, L, Huber, K.V.M, Fedorov, O, Owen, D.R, Londregan, A.T, Bountra, C, Edwards, A, Arrowsmith, C. | | Deposit date: | 2023-06-23 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of PFI-6, a small-molecule chemical probe for the YEATS domain of MLLT1 and MLLT3.
Bioorg.Med.Chem.Lett., 98, 2023
|
|
7PSP
 
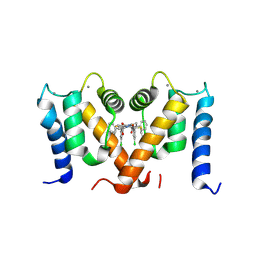 | | Crystal structure of S100A4 labeled with NU000846b. | | Descriptor: | (2R,4R)-1-(2-chloranylethanoyl)-N-(3-chlorophenyl)-4-phenyl-pyrrolidine-2-carboxamide, CALCIUM ION, Protein S100-A4 | | Authors: | Giroud, C, Szommer, T, Coxon, C, Monteiro, O, Christott, T, Bennett, J, Aitmakhanova, K, Raux, B, Newman, J, Elkins, J, Arruda Bezerra, G, Krojer, T, Koekemoer, L, Von Delft, F, Bountr, C, Brennan, P, Fedorov, O. | | Deposit date: | 2021-09-23 | | Release date: | 2022-10-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of S100A4 labeled with NU000846b.
To Be Published
|
|
8C19
 
 | | SARS-CoV-2 NSP3 macrodomain in complex with 1-methyl-4-[5-(morpholin-4-ylcarbonyl)-2-furyl]-1H-pyrrolo[2,3-b]pyridine | | Descriptor: | 1,2-ETHANEDIOL, Non-structural protein 3, [5-(1-methylpyrrolo[2,3-b]pyridin-4-yl)furan-2-yl]-morpholin-4-yl-methanone | | Authors: | Schuller, M, Ahel, I. | | Deposit date: | 2022-12-20 | | Release date: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and Development Strategies for SARS-CoV-2 NSP3 Macrodomain Inhibitors.
Pathogens, 12, 2023
|
|
8C1A
 
 | |
6S1I
 
 | | Crystal Structure of DYRK1A with small molecule inhibitor | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 1A, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Sorrell, F.J, Henderson, S.H, Redondo, C, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Elkins, J.M. | | Deposit date: | 2019-06-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Mining Public Domain Data to Develop Selective DYRK1A Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
6T1I
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with piperazine-urea derivative 1 | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-ethanoylphenyl)-~{N}-[(6-methoxypyridin-3-yl)methyl]piperazine-1-carboxamide, Protein ENL | | Authors: | Chaikuad, A, Heidenreich, D, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
6T1M
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with benzimidazole-amide derivative 4 | | Descriptor: | 1,2-ETHANEDIOL, 4-cyano-~{N}-[2-(piperidin-1-ylmethyl)-1~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Chaikuad, A, Heidenreich, D, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
6T1O
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with benzimidazole-amide derivative 6 | | Descriptor: | 1,2-ETHANEDIOL, 4-iodanyl-~{N}-[2-(piperidin-1-ylmethyl)-3~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Chaikuad, A, Heidenreich, D, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
6T1J
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with piperazine-urea derivative 2 | | Descriptor: | 1,2-ETHANEDIOL, Protein ENL, ~{N}-[[4-(pyrrolidin-1-ylmethyl)phenyl]methyl]-4-thiophen-2-ylcarbonyl-piperazine-1-carboxamide | | Authors: | Chaikuad, A, Heidenreich, D, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
6T1L
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with piperazine-urea derivative 3 | | Descriptor: | 1,2-ETHANEDIOL, Protein ENL, ~{N}-[[4-(diethylaminomethyl)phenyl]methyl]-4-pyrimidin-2-yl-piperazine-1-carboxamide | | Authors: | Chaikuad, A, Heidenreich, D, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
6T1N
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with benzimidazole-amide derivative 5 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-~{N}-[2-(piperidin-1-ylmethyl)-3~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Chaikuad, A, Heidenreich, D, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
7GB5
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDG-MED-0da5ad92-2 (Mpro-x10201) | | Descriptor: | 2-(3-chlorophenyl)-N-(pyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.261 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GB3
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with DAV-CRI-3edb475e-6 (Mpro-x10172) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(1S)-1-(3-chloro-5-fluorophenyl)ethyl]acetamide | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.379 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GGZ
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-3c65e9ce-2 (Mpro-x12719) | | Descriptor: | 2-(4-acetylpiperazin-1-yl)-N-(4-cyclopropylpyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
