3GAR
 
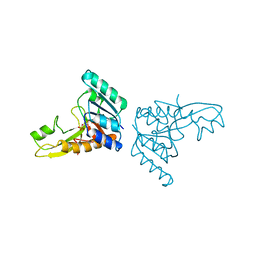 | | A PH-DEPENDENT STABLIZATION OF AN ACTIVE SITE LOOP OBSERVED FROM LOW AND HIGH PH CRYSTAL STRUCTURES OF MUTANT MONOMERIC GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE, PHOSPHATE ION | | Authors: | Su, Y, Yamashita, M.M, Greasley, S.E, Mullen, C.A, Shim, J.H, Jennings, P.A, Benkovic, S.J, Wilson, I.A. | | Deposit date: | 1998-05-13 | | Release date: | 1998-08-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A pH-dependent stabilization of an active site loop observed from low and high pH crystal structures of mutant monomeric glycinamide ribonucleotide transformylase at 1.8 to 1.9 A.
J.Mol.Biol., 281, 1998
|
|
1C3E
 
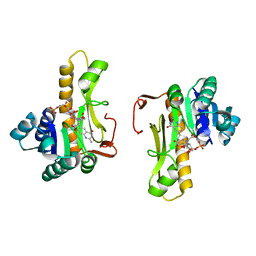 | | NEW INSIGHTS INTO INHIBITOR DESIGN FROM THE CRYSTAL STRUCTURE AND NMR STUDIES OF E. COLI GAR TRANSFORMYLATE IN COMPLEX WITH BETA-GAR AND 10-FORMYL-5,8,10-TRIDEAZAFOLIC ACID. | | Descriptor: | 2-{4-[2-(2-AMINO-4-HYDROXY-QUINAZOLIN-6-YL)-1-CARBOXY-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, GLYCINAMIDE RIBONUCLEOTIDE, GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Authors: | Greasley, S.E, Yamashita, M.M, Cai, H, Benkovic, S.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 1999-07-27 | | Release date: | 1999-12-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New insights into inhibitor design from the crystal structure and NMR studies of Escherichia coli GAR transformylase in complex with beta-GAR and 10-formyl-5,8,10-trideazafolic acid.
Biochemistry, 38, 1999
|
|
1C2T
 
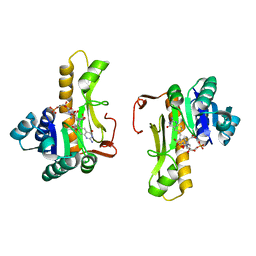 | | NEW INSIGHTS INTO INHIBITOR DESIGN FROM THE CRYSTAL STRUCTURE AND NMR STUDIES OF E. COLI GAR TRANSFORMYLASE IN COMPLEX WITH BETA-GAR AND 10-FORMYL-5,8,10-TRIDEAZAFOLIC ACID. | | Descriptor: | 10-FORMYL-5,8,10-TRIDEAZAFOLIC ACID, GLYCINAMIDE RIBONUCLEOTIDE, GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Authors: | Greasley, S.E, Yamashita, M.M, Cai, H, Benkovic, S.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 1999-07-26 | | Release date: | 2000-01-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New insights into inhibitor design from the crystal structure and NMR studies of Escherichia coli GAR transformylase in complex with beta-GAR and 10-formyl-5,8,10-trideazafolic acid.
Biochemistry, 38, 1999
|
|
2GAR
 
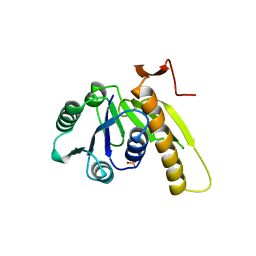 | | A PH-DEPENDENT STABLIZATION OF AN ACTIVE SITE LOOP OBSERVED FROM LOW AND HIGH PH CRYSTAL STRUCTURES OF MUTANT MONOMERIC GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE, PHOSPHATE ION | | Authors: | Su, Y, Yamashita, M.M, Greasley, S.E, Mullen, C.A, Shim, J.H, Jennings, P.A, Benkovic, S.J, Wilson, I.A. | | Deposit date: | 1998-05-13 | | Release date: | 1998-08-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A pH-dependent stabilization of an active site loop observed from low and high pH crystal structures of mutant monomeric glycinamide ribonucleotide transformylase at 1.8 to 1.9 A.
J.Mol.Biol., 281, 1998
|
|
4GH8
 
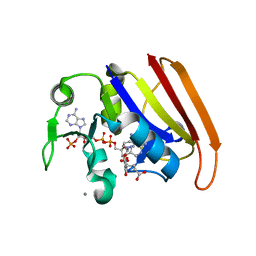 | | Crystal structure of a 'humanized' E. coli dihydrofolate reductase | | Descriptor: | CALCIUM ION, Dihydrofolate reductase, METHOTREXATE, ... | | Authors: | French, J.B, Liu, C.T, Hanoian, P, Pringle, T.H, Hammes-Schiffer, S, Benkovic, S.J. | | Deposit date: | 2012-08-07 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Functional significance of evolving protein sequence in dihydrofolate reductase from bacteria to humans.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2V0G
 
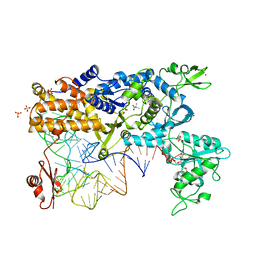 | | LEUCYL-TRNA SYNTHETASE FROM THERMUS THERMOPHILUS COMPLEXED WITH A tRNA(leu) transcript with 5-FLUORO-1,3-DIHYDRO-1-HYDROXY-2,1- BENZOXABOROLE (AN2690) forming an adduct to the ribose of adenosine- 76 in the enzyme editing site. | | Descriptor: | AMINOACYL-TRNA SYNTHETASE, LEUCINE, MERCURY (II) ION, ... | | Authors: | Rock, F, Mao, W, Yaremchuk, A, Tukalo, M, Crepin, T, Zhou, H, Zhang, Y, Hernandez, V, Akama, T, Baker, S, Plattner, J, Shapiro, L, Martinis, S.A, Benkovic, S.J, Cusack, S, Alley, M.R.K. | | Deposit date: | 2007-05-14 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | An Antifungal Agent Inhibits an Aminoacyl-tRNA Synthetase by Trapping tRNA in the Editing Site.
Science, 316, 2007
|
|
2V0C
 
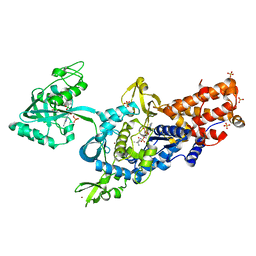 | | LEUCYL-TRNA SYNTHETASE FROM THERMUS THERMOPHILUS COMPLEXED WITH A SULPHAMOYL ANALOGUE OF LEUCYL-ADENYLATE In the synthetic site and an adduct of AMP with 5-Fluoro-1,3-dihydro-1-hydroxy-2,1-benzoxaborole (AN2690) in the editing site | | Descriptor: | AMINOACYL-TRNA SYNTHETASE, LEUCINE, SULFATE ION, ... | | Authors: | Rock, F, Mao, W, Yaremchuk, A, Tukalo, M, Crepin, T, Zhou, H, Zhang, Y, Hernandez, V, Akama, T, Baker, S, Plattner, J, Shapiro, L, Martinis, S.A, Benkovic, S.J, Cusack, S, Alley, M.R.K. | | Deposit date: | 2007-05-14 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An Antifungal Agent Inhibits an Aminoacyl-tRNA Synthetase by Trapping tRNA in the Editing Site.
Science, 316, 2007
|
|
4P66
 
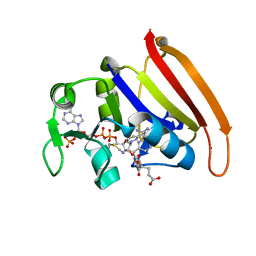 | | Electrostatics of Active Site Microenvironments of E. coli DHFR | | Descriptor: | CALCIUM ION, Dihydrofolate reductase, METHOTREXATE, ... | | Authors: | Liu, C.T, Layfield, J.P, Stewart III, R.J, French, J.B, Hanoian, P, Asbury, J.B, Hammes-Schiffer, S, Benkovic, S.J. | | Deposit date: | 2014-03-21 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Probing the electrostatics of active site microenvironments along the catalytic cycle for Escherichia coli dihydrofolate reductase.
J.Am.Chem.Soc., 136, 2014
|
|
4P68
 
 | | Electrostatics of Active Site Microenvironments for E. coli DHFR | | Descriptor: | ACETATE ION, CALCIUM ION, Dihydrofolate reductase, ... | | Authors: | Liu, C.T, Layfield, J.P, Stewart III, R.J, French, J.B, Hanoian, P, Asbury, J.B, Hammes-Schiffer, S, Benkovic, S.J. | | Deposit date: | 2014-03-22 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Probing the electrostatics of active site microenvironments along the catalytic cycle for Escherichia coli dihydrofolate reductase.
J.Am.Chem.Soc., 136, 2014
|
|
1OZ0
 
 | | CRYSTAL STRUCTURE OF THE HOMODIMERIC BIFUNCTIONAL TRANSFORMYLASE AND CYCLOHYDROLASE ENZYME AVIAN ATIC IN COMPLEX WITH A MULTISUBSTRATE ADDUCT INHIBITOR BETA-DADF. | | Descriptor: | 2-[4-((2-AMINO-4-OXO-3,4-DIHYDRO-PYRIDO[3,2-D]PYRIMIDIN-6-YLMETHYL)-{3-[5-CARBAMOYL-3-(3,4- DIHYDROXY-5-PHOSPHONOOXYMETHYL-TETRAHYDRO-FURAN-2-YL)-3H-IMIDAZOL-4-YL]-ACRYLOYL}-AMINO)-BENZOYLAMINO]- PENTANEDIOIC ACID, Bifunctional purine biosynthesis protein PURH, PHOSPHATE ION, ... | | Authors: | Wolan, D.W, Greasley, S.E, Wall, M.J, Benkovic, S.J, Wilson, I.A. | | Deposit date: | 2003-04-07 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Avian AICAR Transformylase with a Multisubstrate Adduct Inhibitor beta-DADF Identifies the Folate Binding Site.
Biochemistry, 42, 2003
|
|
1JKX
 
 | | Unexpected formation of an epoxide-derived multisubstrate adduct inhibitor on the active site of GAR transformylase | | Descriptor: | N-[5'-O-PHOSPHONO-RIBOFURANOSYL]-2-[2-HYDROXY-2-[4-[GLUTAMIC ACID]-N-CARBONYLPHENYL]-3-[2-AMINO-4-HYDROXY-QUINAZOLIN-6-YL]-PROPANYLAMINO]-ACETAMIDE, PHOSPHORIBOSYLGLYCINAMIDE FORMYLTRANSFERASE | | Authors: | Greasley, S.E, Marsilje, T.H, Cai, H, Baker, S, Benkovic, S.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2001-07-13 | | Release date: | 2001-11-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unexpected formation of an epoxide-derived multisubstrate adduct inhibitor on the active site of GAR transformylase.
Biochemistry, 40, 2001
|
|
1EZ1
 
 | | STRUCTURE OF ESCHERICHIA COLI PURT-ENCODED GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE COMPLEXED WITH MG, AMPPNP, AND GAR | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ACETATE ION, GLYCINAMIDE RIBONUCLEOTIDE, ... | | Authors: | Thoden, J.B, Firestine, S, Nixon, A, Benkovic, S.J, Holden, H.M. | | Deposit date: | 2000-05-09 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular structure of Escherichia coli PurT-encoded glycinamide ribonucleotide transformylase.
Biochemistry, 39, 2000
|
|
1EYZ
 
 | | STRUCTURE OF ESCHERICHIA COLI PURT-ENCODED GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE COMPLEXED WITH MG AND AMPPNP | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Thoden, J.B, Firestine, S, Nixon, A, Benkovic, S.J, Holden, H.M. | | Deposit date: | 2000-05-09 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular structure of Escherichia coli PurT-encoded glycinamide ribonucleotide transformylase.
Biochemistry, 39, 2000
|
|
1G8M
 
 | | CRYSTAL STRUCTURE OF AVIAN ATIC, A BIFUNCTIONAL TRANSFORMYLASE AND CYCLOHYDROLASE ENZYME IN PURINE BIOSYNTHESIS AT 1.75 ANG. RESOLUTION | | Descriptor: | AICAR TRANSFORMYLASE-IMP CYCLOHYDROLASE, GUANOSINE-5'-MONOPHOSPHATE, POTASSIUM ION | | Authors: | Greasley, S.E, Horton, P, Beardsley, G.P, Benkovic, S.J, Wilson, I.A. | | Deposit date: | 2000-11-17 | | Release date: | 2001-04-27 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a bifunctional transformylase and cyclohydrolase enzyme in purine biosynthesis.
Nat.Struct.Biol., 8, 2001
|
|
1KJQ
 
 | | Crystal structure of glycinamide ribonucleotide transformylase in complex with Mg-ADP | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Firestine, S.M, Benkovic, S.J, Holden, H.M. | | Deposit date: | 2001-12-05 | | Release date: | 2002-06-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | PurT-encoded glycinamide ribonucleotide transformylase. Accommodation of adenosine nucleotide analogs within the active site.
J.Biol.Chem., 277, 2002
|
|
1KJJ
 
 | | Crystal structure of glycniamide ribonucleotide transformylase in complex with Mg-ATP-gamma-S | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Thoden, J.B, Firestine, S.M, Benkovic, S.J, Holden, H.M. | | Deposit date: | 2001-12-04 | | Release date: | 2002-06-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | PurT-encoded glycinamide ribonucleotide transformylase. Accommodation of adenosine nucleotide analogs within the active site.
J.Biol.Chem., 277, 2002
|
|
1KJI
 
 | | Crystal structure of glycinamide ribonucleotide transformylase in complex with Mg-AMPPCP | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Thoden, J.B, Firestine, S.M, Benkovic, S.J, Holden, H.M. | | Deposit date: | 2001-12-04 | | Release date: | 2002-06-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | PurT-encoded glycinamide ribonucleotide transformylase. Accommodation of adenosine nucleotide analogs within the active site.
J.Biol.Chem., 277, 2002
|
|
1KJ9
 
 | | Crystal structure of purt-encoded glycinamide ribonucleotide transformylase complexed with Mg-ATP | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Thoden, J.B, Firestine, S.M, Benkovic, S.J, Holden, H.M. | | Deposit date: | 2001-12-04 | | Release date: | 2002-06-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | PurT-encoded glycinamide ribonucleotide transformylase. Accommodation of adenosine nucleotide analogs within the active site.
J.Biol.Chem., 277, 2002
|
|
1KJ8
 
 | | Crystal Structure of PurT-Encoded Glycinamide Ribonucleotide Transformylase in Complex with Mg-ATP and GAR | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Thoden, J.B, Firestine, S.M, Benkovic, S.J, Holden, H.M. | | Deposit date: | 2001-12-04 | | Release date: | 2002-06-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | PurT-encoded glycinamide ribonucleotide transformylase. Accommodation of adenosine nucleotide analogs within the active site.
J.Biol.Chem., 277, 2002
|
|
1NJS
 
 | | human GAR Tfase in complex with hydrolyzed form of 10-trifluoroacetyl-5,10-dideaza-acyclic-5,6,7,8-tetrahydrofolic acid | | Descriptor: | N-{4-[(1R)-4-[(2R,4R,5S)-2,4-DIAMINO-6-OXOHEXAHYDROPYRIMIDIN-5-YL]-1-(2,2,2-TRIFLUORO-1,1-DIHYDROXYETHYL)BUTYL]BENZOYL}-D-GLUTAMIC ACID, PHOSPHATE ION, Phosphoribosylglycinamide formyltransferase | | Authors: | Zhang, Y, Desharnais, J, Marsilje, T.H, Li, C, Hedrick, M.P, Gooljarsingh, L.T, Tavassoli, A, Benkovic, S.J, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2003-01-02 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Rational Design, Synthesis, Evaluation, and Crystal Structure of a Potent Inhibitor of Human GAR Tfase: 10-(Trifluoroacetyl)-5,10-dideazaacyclic-5,6,7,8-tetrahydrofolic Acid
Biochemistry, 42, 2003
|
|
43CA
 
 | |
43C9
 
 | |
7DFR
 
 | |
3IX9
 
 | |
6DFR
 
 | |
