1ANU
 
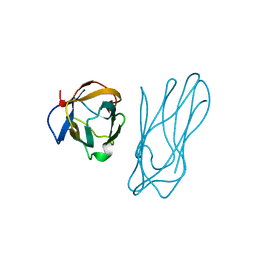 | | COHESIN-2 DOMAIN OF THE CELLULOSOME FROM CLOSTRIDIUM THERMOCELLUM | | Descriptor: | COHESIN-2 | | Authors: | Shimon, L.J.W, Yaron, S, Shoham, Y, Lamed, R, Morag, E, Bayer, E.A, Frolow, F. | | Deposit date: | 1996-07-19 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A cohesin domain from Clostridium thermocellum: the crystal structure provides new insights into cellulosome assembly.
Structure, 5, 1997
|
|
4WKZ
 
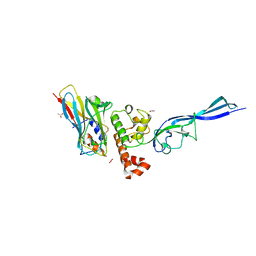 | | Complex of autonomous ScaG cohesin CohG and X-doc domains | | Descriptor: | ACETATE ION, Autonomous cohesin, CALCIUM ION, ... | | Authors: | Voronov-Goldman, M, Lamed, R, Bayer, E.A, Yaniv, O, Shimon, L.J.W. | | Deposit date: | 2014-10-05 | | Release date: | 2015-12-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Complex of autonomous ScaG cohesin CohG and X-doc domains
To Be Published
|
|
3L8Q
 
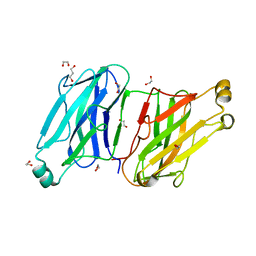 | | Structure analysis of the type II cohesin dyad from the adaptor ScaA scaffoldin of Acetivibrio cellulolyticus | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, Cellulosomal scaffoldin adaptor protein B, ... | | Authors: | Noach, I, Frolow, F, Bayer, E.A. | | Deposit date: | 2010-01-03 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Modular Arrangement of a Cellulosomal Scaffoldin Subunit Revealed from the Crystal Structure of a Cohesin Dyad
J.Mol.Biol., 399, 2010
|
|
3KCP
 
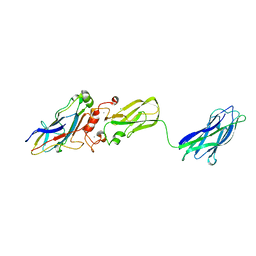 | | Crystal structure of interacting Clostridium thermocellum multimodular components | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cellulosomal-scaffolding protein A, ... | | Authors: | Adams, J.J, Currie, M.A, Bayer, E.A, Jia, Z, Smith, S.P. | | Deposit date: | 2009-10-21 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Insights into Higher-Order Organization of the Cellulosome Revealed by a Dissect-and-Build Approach: Crystal Structure of Interacting Clostridium thermocellum Multimodular Components
J.Mol.Biol., 396, 2010
|
|
2WO4
 
 | | 3b' carbohydrate-binding module from the Cel9V glycoside hydrolase from Clostridium thermocellum, in-house data | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCOSIDE HYDROLASE, ... | | Authors: | Petkun, S, Jindou, S, Shimon, L.J.W, Bayer, E.A, Lamed, R, Frolow, F. | | Deposit date: | 2009-07-21 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a Family 3B' Carbohydrate-Binding Module from the Cel9V Glycoside Hydrolase from Clostridium Thermocellum: Structural Diversity and Implications for Carbohydrate Binding
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2WOB
 
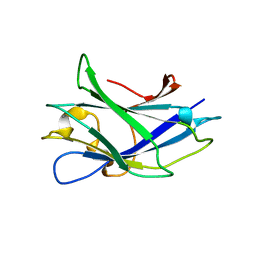 | | 3b' carbohydrate-binding module from the Cel9V glycoside hydrolase from Clostridium thermocellum. Orthorhombic structure | | Descriptor: | CALCIUM ION, GLYCOSIDE HYDROLASE, FAMILY 9 | | Authors: | Petkun, S, Jindou, S, Shimon, L.J.W, Bayer, E.A, Lamed, R, Frolow, F. | | Deposit date: | 2009-07-22 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a Family 3B' Carbohydrate-Binding Module from the Cel9V Glycoside Hydrolase from Clostridium Thermocellum: Structural Diversity and Implications for Carbohydrate Binding
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2WNX
 
 | | 3b' carbohydrate-binding module from the Cel9V glycoside hydrolase from Clostridium thermocellum | | Descriptor: | CALCIUM ION, FORMIC ACID, GLYCOSIDE HYDROLASE, ... | | Authors: | Petkun, S, Jindou, S, Shimon, L.J.W, Bayer, E.A, Lamed, R, Frolow, F. | | Deposit date: | 2009-07-20 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structure of a Family 3B' Carbohydrate-Binding Module from the Cel9V Glycoside Hydrolase from Clostridium Thermocellum: Structural Diversity and Implications for Carbohydrate Binding
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2XDH
 
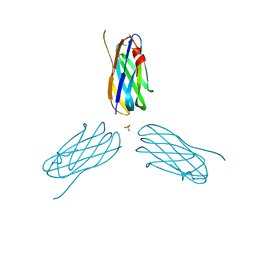 | | Non-cellulosomal cohesin from the hyperthermophilic archaeon Archaeoglobus fulgidus | | Descriptor: | CHLORIDE ION, COHESIN, MAGNESIUM ION, ... | | Authors: | Voronov-Goldman, M, Lamed, R, Noach, I, Borovok, I, Kwiat, M, Rosenheck, S, Shimon, L.J.W, Bayer, E.A, Frolow, F. | | Deposit date: | 2010-05-02 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Non-Cellulosomal Cohesin from the Hyperthermophilic Archaeon Archaeoglobus Fulgidus
Proteins, 79, 2011
|
|
3FNK
 
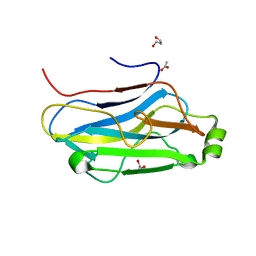 | | Crystal structure of the second type II cohesin module from the cellulosomal adaptor ScaA scaffoldin of Acetivibrio cellulolyticus | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 1,4-BUTANEDIOL, ... | | Authors: | Noach, I, Frolow, F, Bayer, E.A. | | Deposit date: | 2008-12-25 | | Release date: | 2009-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Intermodular Linker Flexibility Revealed from Crystal Structures of Adjacent Cellulosomal Cohesins of Acetivibrio cellulolyticus
J.Mol.Biol., 391, 2009
|
|
3GHP
 
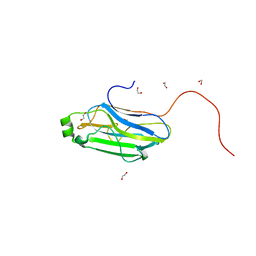 | |
4JO5
 
 | | CBM3a-L domain with flanking linkers from scaffoldin cipA of cellulosome of Clostridium thermocellum | | Descriptor: | 1-METHYLIMIDAZOLE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Shimon, L.J.W, Frolow, F, Bayer, E.A, Yaniv, O, Lamed, R, Morag, E. | | Deposit date: | 2013-03-16 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of a family 3a carbohydrate-binding module from the cellulosomal scaffoldin CipA of Clostridium thermocellum with flanking linkers: implications for cellulosome structure.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
6QDI
 
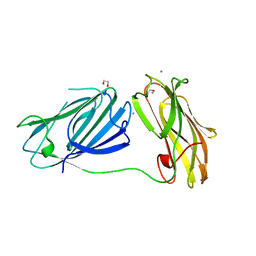 | | anti-sigma factor domain-containing protein from Clostridium clariflavum | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PA14 domain-containing protein | | Authors: | Voronov, M, Bayer, E.A, Livnah, O. | | Deposit date: | 2019-01-01 | | Release date: | 2019-06-12 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Distinctive ligand-binding specificities of tandem PA14 biomass-sensory elements from Clostridium thermocellum and Clostridium clariflavum.
Proteins, 87, 2019
|
|
6QE7
 
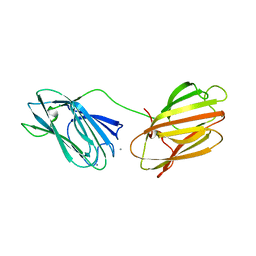 | | anti-sigma factor domain-containing protein | | Descriptor: | Anti-sigma-I factor RsgI3, CALCIUM ION | | Authors: | Voronov, M, Livnah, O, Bayer, E.A. | | Deposit date: | 2019-01-07 | | Release date: | 2019-06-12 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Distinctive ligand-binding specificities of tandem PA14 biomass-sensory elements from Clostridium thermocellum and Clostridium clariflavum.
Proteins, 87, 2019
|
|
2VO8
 
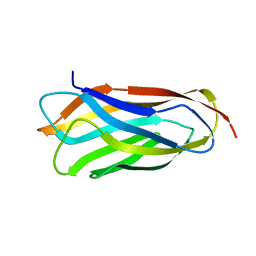 | | Cohesin module from Clostridium perfringens ATCC13124 family 33 glycoside hydrolase. | | Descriptor: | EXO-ALPHA-SIALIDASE | | Authors: | Gregg, K, Adams, J.J, Bayer, E.A, Boraston, A.B, Smith, S.P. | | Deposit date: | 2008-02-08 | | Release date: | 2008-09-02 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Clostridium Perfringens Toxin Complex Formation.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2XFG
 
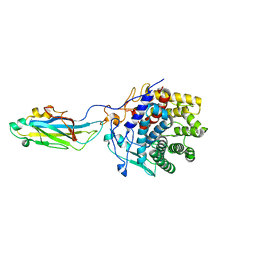 | | Reassembly and co-crystallization of a family 9 processive endoglucanase from separately expressed GH9 and CBM3c modules | | Descriptor: | CALCIUM ION, CHLORIDE ION, ENDOGLUCANASE 1 | | Authors: | Petkun, S, Lamed, R, Jindou, S, Burstein, T, Yaniv, O, Shoham, Y, Shimon, J.W.L, Bayer, E.A, Frolow, F. | | Deposit date: | 2010-05-24 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.679 Å) | | Cite: | Reassembly and Co-Crystallization of a Family 9 Processive Endoglucanase from its Component Parts: Structural and Functional Significance of Intermodular Linker
Peerj, 3, 2015
|
|
2XQO
 
 | | CtCel124: a cellulase from Clostridium thermocellum | | Descriptor: | Dockerin type 1, NICKEL (II) ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Carvalho, A.L, Verze, G, Bras, J.L.A, Cartmell, A, Bayer, E.A, Vazana, Y, Correia, M.A.S, Prates, J.A.M, Gilbert, H.J, Fontes, C.M.G.A, Romao, M.J. | | Deposit date: | 2010-09-06 | | Release date: | 2011-03-02 | | Last modified: | 2021-02-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Insights Into a Unique Cellulase Fold and Mechanism of Cellulose Hydrolysis
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2XBT
 
 | | Structure of a scaffoldin carbohydrate-binding module family 3b from the cellulosome of Bacteroides cellulosolvens: Structural diversity and implications for carbohydrate binding | | Descriptor: | CELLULOSOMAL SCAFFOLDIN, NITRATE ION | | Authors: | Yaniv, O, Shimon, L.J.W, Bayer, E.A, Lamed, R, Frolow, F. | | Deposit date: | 2010-04-15 | | Release date: | 2011-04-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.832 Å) | | Cite: | Scaffoldin-Borne Family 3B Carbohydrate-Binding Module from the Cellulosome of Bacteroides Cellulosolvens: Structural Diversity and Significance of Calcium for Carbohydrate Binding
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2YLK
 
 | | Carbohydrate-binding module CBM3b from the cellulosomal cellobiohydrolase 9A from Clostridium thermocellum | | Descriptor: | CELLULOSE 1,4-BETA-CELLOBIOSIDASE | | Authors: | Yaniv, O, Petkun, S, Shimon, L.J.W, Bayer, E.A, Lamed, R, Frolow, F. | | Deposit date: | 2011-06-02 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Single Mutation Reforms the Binding Activity of an Adhesion-Deficient Family 3 Carbohydrate-Binding Module
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1G43
 
 | | CRYSTAL STRUCTURE OF A FAMILY IIIA CBD FROM CLOSTRIDIUM CELLULOLYTICUM | | Descriptor: | CALCIUM ION, SCAFFOLDING PROTEIN, ZINC ION | | Authors: | Shimon, L.J.W, Pages, S, Belaich, A, Belaich, J.-P, Bayer, E.A, Lamed, R, Shoham, Y, Frolow, F. | | Deposit date: | 2000-10-26 | | Release date: | 2000-12-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a family IIIa scaffoldin CBD from the cellulosome of Clostridium cellulolyticum at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
2JNK
 
 | |
2JH2
 
 | | X-ray crystal structure of a cohesin-like module from Clostridium perfringens | | Descriptor: | O-GLCNACASE NAGJ | | Authors: | Chitayat, S, Gregg, K, Adams, J.J, Ficko-Blean, E, Bayer, E.A, Boraston, A.B, Smith, S.P. | | Deposit date: | 2007-02-19 | | Release date: | 2007-11-06 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-Dimensional Structure of a Putative Non- Cellulosomal Cohesin Module from a Clostridium Perfringens Family 84 Glycoside Hydrolase.
J.Mol.Biol., 375, 2008
|
|
1TYJ
 
 | | Crystal Structure Analysis of type II Cohesin A11 from Bacteroides cellulosolvens | | Descriptor: | 1,2-ETHANEDIOL, METHANOL, cellulosomal scaffoldin | | Authors: | Noach, I, Frolow, F, Jakoby, H, Rosenheck, S, Shimon, L.J.W, Lamed, R, Bayer, E.A. | | Deposit date: | 2004-07-08 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a type-II cohesin module from the Bacteroides cellulosolvens cellulosome reveals novel and distinctive secondary structural elements
J.Mol.Biol., 348, 2005
|
|
2VN6
 
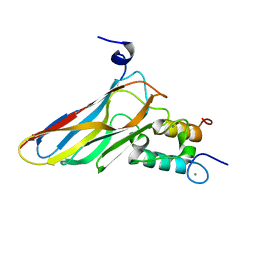 | | The Clostridium cellulolyticum dockerin displays a dual binding mode for its cohesin partner | | Descriptor: | CALCIUM ION, ENDOGLUCANASE A, SCAFFOLDING PROTEIN | | Authors: | Pinheiro, B.A, Prates, J.A.M, Proctor, M.R, Gilbert, H.J, Davies, G.J, Money, V.A, Martinez-Fleites, C, Bayer, E.A, Fontes, C.M.G.A, Fierobe, H.P. | | Deposit date: | 2008-01-31 | | Release date: | 2008-05-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | The Clostridium Cellulolyticum Dockerin Displays a Dual Binding Mode for its Cohesin Partner.
J.Biol.Chem., 283, 2008
|
|
2VN5
 
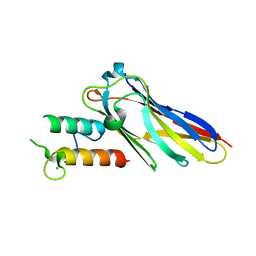 | | The Clostridium cellulolyticum dockerin displays a dual binding mode for its cohesin partner | | Descriptor: | CALCIUM ION, ENDOGLUCANASE A, SCAFFOLDING PROTEIN | | Authors: | Pinheiro, B.A, Prates, J.A.M, Proctor, M.R, Gilbert, H.J, Davies, G.J, Money, V.A, Martinez-Fleites, C, Bayer, E.A, Fontes, C.M.G.A, Fierobe, H.P. | | Deposit date: | 2008-01-31 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Clostridium Cellulolyticum Dockerin Displays a Dual Binding Mode for its Cohesin Partner.
J.Biol.Chem., 283, 2008
|
|
3ZQX
 
 | | Carbohydrate-binding module CBM3b from the cellulosomal cellobiohydrolase 9A from Clostridium thermocellum | | Descriptor: | CALCIUM ION, CELLULOSE 1,4-BETA-CELLOBIOSIDASE | | Authors: | Yaniv, O, Petkun, S, Shimon, L.J.W, Bayer, E.A, Lamed, R, Frolow, F. | | Deposit date: | 2011-06-12 | | Release date: | 2012-04-25 | | Last modified: | 2012-07-11 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | A Single Mutation Reforms the Binding Activity of an Adhesion-Deficient Family 3 Carbohydrate-Binding Module
Acta Crystallogr.,Sect.D, 68, 2012
|
|
