3ZRS
 
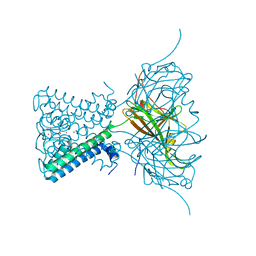 | | X-ray crystal structure of a KirBac potassium channel highlights a mechanism of channel opening at the bundle-crossing gate. | | Descriptor: | ATP-SENSITIVE INWARD RECTIFIER POTASSIUM CHANNEL 10, CHLORIDE ION, POTASSIUM ION | | Authors: | Bavro, V.N, De Zorzi, R, Schmidt, M.R, Muniz, J.R.C, Zubcevic, L, Sansom, M.S.P, Venien-Bryan, C, Tucker, S.J. | | Deposit date: | 2011-06-17 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure of a Kirbac Potassium Channel with an Open Bundle Crossing Indicates a Mechanism of Channel Gating
Nat.Struct.Mol.Biol., 19, 2012
|
|
2VDE
 
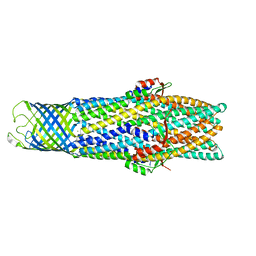 | | Crystal Structure of the Open State of TolC Outer Membrane Component of Mutlidrug Efflux Pumps | | Descriptor: | CHLORIDE ION, OUTER MEMBRANE PROTEIN TOLC | | Authors: | Bavro, V.N, Pietras, Z, Furnham, N, Perez-Cano, L, Fernandez-Recio, J, Pei, X.Y, Truer, R, Misra, R, Luisi, B. | | Deposit date: | 2007-10-04 | | Release date: | 2008-04-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Assembly and Channel Opening in a Bacterial Drug Efflux Machine.
Mol.Cell, 30, 2008
|
|
2VDD
 
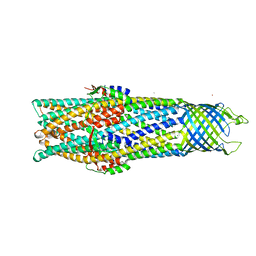 | | Crystal Structure of the Open State of TolC Outer Membrane Component of Mutlidrug Efflux Pumps | | Descriptor: | CHLORIDE ION, OUTER MEMBRANE PROTEIN TOLC, POTASSIUM ION | | Authors: | Bavro, V.N, Pietras, Z, Furnham, N, Perez-Cano, L, Fernandez-Recio, J, Pei, X.Y, Truer, R, Misra, R, Luisi, B. | | Deposit date: | 2007-10-04 | | Release date: | 2008-04-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Assembly and Channel Opening in a Bacterial Drug Efflux Machine.
Mol.Cell, 30, 2008
|
|
1KJT
 
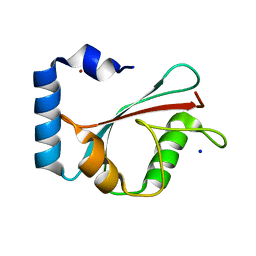 | | Crystal Structure of the GABA(A) Receptor Associated Protein, GABARAP | | Descriptor: | GABARAP, NICKEL (II) ION, SODIUM ION | | Authors: | Bavro, V.N, Sola, M, Bracher, A, Kneussel, M, Betz, H, Weissenhorn, W. | | Deposit date: | 2001-12-05 | | Release date: | 2002-04-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the GABA(A)-receptor-associated protein, GABARAP.
EMBO Rep., 3, 2002
|
|
1T3E
 
 | | Structural basis of dynamic glycine receptor clustering | | Descriptor: | 49-mer fragment of Glycine receptor beta chain, Gephyrin, SULFATE ION | | Authors: | Sola, M, Bavro, V.N, Timmins, J, Franz, T, Ricard-Blum, S, Schoehn, G, Ruigrok, R.W.H, Paarmann, I, Saiyed, T, O'Sullivan, G.A. | | Deposit date: | 2004-04-26 | | Release date: | 2004-07-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural basis of dynamic glycine receptor clustering by gephyrin
Embo J., 23, 2004
|
|
7A2D
 
 | | Structure-function analyses of dual-BON domain protein DolP identifies phospholipid binding as a new mechanism for protein localisation to the cell division site | | Descriptor: | Uncharacterized protein YraP | | Authors: | Bryant, J.A, Morris, F.C, Knowles, T.J, Maderbocus, R, Heinz, E, Boelter, G, Alodaini, D, Colyer, A, Wotherspoon, P.J, Staunton, K.A, Jeeves, M, Browning, D.F, Sevastsyanovich, Y.R, Wells, T.J, Rossiter, A.E, Bavro, V.N, Sridhar, P, Ward, D.G, Chong, Z.S, Goodall, E.C.A, Icke, C, Teo, A, Chng, S.S, Roper, D.I, Lithgow, T, Cunningham, A.F, Banzhaf, M, Overduin, M, Henderson, I.R. | | Deposit date: | 2020-08-17 | | Release date: | 2020-12-30 | | Last modified: | 2021-02-10 | | Method: | SOLUTION NMR | | Cite: | Structure of dual BON-domain protein DolP identifies phospholipid binding as a new mechanism for protein localisation.
Elife, 9, 2020
|
|
4LP8
 
 | | A Novel Open-State Crystal Structure of the Prokaryotic Inward Rectifier KirBac3.1 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Inward rectifier potassium channel Kirbac3.1, ... | | Authors: | Zubcevic, L, Bavro, V.N, Muniz, J.R.C, Schmidt, M.R, Wang, S, De Zorzi, R, Venien-Bryan, C, Sansom, M.S.P, Nichols, C.G, Tucker, S.J. | | Deposit date: | 2013-07-15 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Control of KirBac3.1 Potassium Channel Gating at the Interface between Cytoplasmic Domains.
J.Biol.Chem., 289, 2014
|
|
6EI0
 
 | | Cytosolic copper storage protein Csp from Streptomyces lividans: apo form | | Descriptor: | Cytosolic copper storage protein (Ccsp), GLYCEROL, SULFATE ION, ... | | Authors: | Straw, M.L, Chaplin, A.K, Hough, M.A, Worrall, J.A.R. | | Deposit date: | 2017-09-15 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | A cytosolic copper storage protein provides a second level of copper tolerance in Streptomyces lividans.
Metallomics, 10, 2018
|
|
6EK9
 
 | | Cytosolic copper storage protein Csp from Streptomyces lividans: Cu loaded form | | Descriptor: | COPPER (I) ION, Cytosolic copper storage protein | | Authors: | Straw, M.L, Chaplin, A.K, Hough, M.A, Worrall, J.A.R. | | Deposit date: | 2017-09-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A cytosolic copper storage protein provides a second level of copper tolerance in Streptomyces lividans.
Metallomics, 10, 2018
|
|
4USC
 
 | | Crystal structure of peroxidase from palm tree Chamaerops excelsa | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Bernardes, A, Santos, J.C, Textor, L.C, Cuadrado, N.H, Kostetsky, E.Y, Roig, M.G, Muniz, J.R.C, Shnyrov, V.L, Polikarpov, I. | | Deposit date: | 2014-07-07 | | Release date: | 2015-05-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure Analysis of Peroxidase from the Palm Tree Chamaerops Excelsa.
Biochimie, 111, 2015
|
|
6Z12
 
 | | Salmonella AcrB solubilised in the SMA copolymer | | Descriptor: | Efflux pump membrane transporter | | Authors: | Muench, s.p, Johnson, R.M. | | Deposit date: | 2020-05-11 | | Release date: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM Structure and Molecular Dynamics Analysis of the Fluoroquinolone Resistant Mutant of the AcrB Transporter fromSalmonella.
Microorganisms, 8, 2020
|
|
