4RUK
 
 | |
3X1J
 
 | |
3X1M
 
 | |
3X1K
 
 | |
4KY9
 
 | |
5NGM
 
 | | 2.9S structure of the 70S ribosome composing the S. aureus 100S complex | | Descriptor: | 16S ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Matzov, D, Aibara, S, Zimmerman, E, Bashan, A, Amunts, A, Yonath, A. | | Deposit date: | 2017-03-18 | | Release date: | 2017-10-04 | | Last modified: | 2018-11-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The cryo-EM structure of hibernating 100S ribosome dimer from pathogenic Staphylococcus aureus.
Nat Commun, 8, 2017
|
|
6FXC
 
 | | The cryo-EM structure of hibernating 100S ribosome dimer from pathogenic Staphylococcus aureus | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Matzov, D, Aibara, S, Zimmerman, E, Bashan, A, Kidmose, R, Amunts, A, Yonath, A. | | Deposit date: | 2018-03-08 | | Release date: | 2018-03-21 | | Last modified: | 2018-11-21 | | Method: | ELECTRON MICROSCOPY (6.76 Å) | | Cite: | The cryo-EM structure of hibernating 100S ribosome dimer from pathogenic Staphylococcus aureus.
Nat Commun, 8, 2017
|
|
2LFA
 
 | | Oligonucleotide duplex contaning (5'S)-8,5'-cyclo-2'-deoxyguansine | | Descriptor: | DNA (5'-D(*AP*CP*AP*AP*AP*CP*AP*CP*GP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*CP*(2LF)P*TP*GP*TP*TP*TP*GP*T)-3') | | Authors: | Huang, H, Das, R.S, Basu, A, Stone, M.P. | | Deposit date: | 2011-06-29 | | Release date: | 2012-01-04 | | Method: | SOLUTION NMR | | Cite: | Structure of (5'S)-8,5'-Cyclo-2'-deoxyguanosine in DNA.
J.Am.Chem.Soc., 133, 2011
|
|
2LFX
 
 | | Structure of the duplex when (5'S)-8,5'-cyclo-2'-deoxyguanosine is placed opposite dT | | Descriptor: | DNA (5'-D(*AP*CP*AP*AP*AP*CP*AP*TP*GP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*CP*(2LF)P*TP*GP*TP*TP*TP*GP*T)-3') | | Authors: | Huang, H, Das, R.S, Basu, A, Stone, M.P. | | Deposit date: | 2011-07-18 | | Release date: | 2012-06-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structures of (5'S)-8,5'-Cyclo-2'-deoxyguanosine Mismatched with dA or dT.
Chem.Res.Toxicol., 25, 2012
|
|
2LG0
 
 | | structure of the duplex containing (5'S)-8,5'-cyclo-2'-deoxyadenosine | | Descriptor: | DNA (5'-D(*AP*CP*AP*AP*AP*CP*AP*TP*GP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*CP*(02I)P*TP*GP*TP*TP*TP*GP*T)-3') | | Authors: | Huang, H, Das, R.S, Basu, A, Stone, M.P. | | Deposit date: | 2011-07-18 | | Release date: | 2012-06-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of (5'S)-8,5'-cyclo-2'-deoxyguanosine in DNA.
J.Am.Chem.Soc., 133, 2011
|
|
2LFY
 
 | | Structure of the duplex when (5'S)-8,5'-cyclo-2'-deoxyguanosine is placed opposite dA | | Descriptor: | DNA (5'-D(*AP*CP*AP*AP*AP*CP*AP*AP*GP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*CP*(2LF)P*TP*GP*TP*TP*TP*GP*T)-3') | | Authors: | Huang, H, Das, R.S, Basu, A, Stone, M.P. | | Deposit date: | 2011-07-18 | | Release date: | 2012-06-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structures of (5'S)-8,5'-Cyclo-2'-deoxyguanosine Mismatched with dA or dT.
Chem.Res.Toxicol., 25, 2012
|
|
4FBU
 
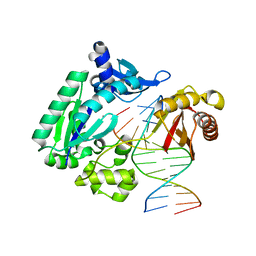 | | Dpo4 polymerase pre-insertion binary complex with the N-(deoxyguanosin-8-yl)-1-aminopyrene lesion | | Descriptor: | CALCIUM ION, DNA polymerase IV, DNA primer, ... | | Authors: | Kirouac, K, Basu, A, Ling, H. | | Deposit date: | 2012-05-23 | | Release date: | 2013-06-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural mechanism of replication stalling on a bulky amino-polycyclic aromatic hydrocarbon DNA adduct by a y family DNA polymerase.
J.Mol.Biol., 425, 2013
|
|
