6VR3
 
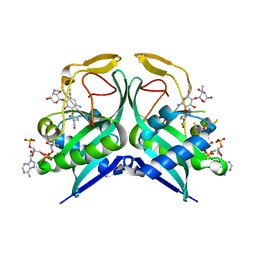 | | Aminoglycoside N-2'-Acetyltransferase-Ia [AAC(2')-Ia] in complex with acetylated-netilmicin and CoA | | Descriptor: | Aminoglycoside 2'-N-acetyltransferase, COENZYME A, N-[(2S,3R)-2-{[(1R,2S,3S,4R,6S)-6-amino-3-{[3-deoxy-4-C-methyl-3-(methylamino)-beta-L-lyxopyranosyl]oxy}-4-(ethylamino) -2-hydroxycyclohexyl]oxy}-6-(aminomethyl)-3,4-dihydro-2H-pyran-3-yl]acetamide | | Authors: | Bassenden, A.V, Berghuis, A.M. | | Deposit date: | 2020-02-06 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and phylogenetic analyses of resistance to next-generation aminoglycosides conferred by AAC(2') enzymes.
Sci Rep, 11, 2021
|
|
6VR2
 
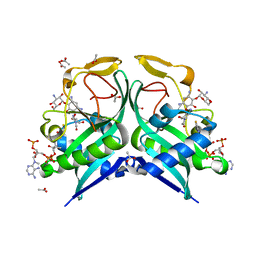 | | Aminoglycoside N-2'-Acetyltransferase-Ia [AAC(2')-Ia] in complex with acetylated-tobramycin and CoA | | Descriptor: | (1S,2S,3R,4S,6R)-3-{[2-(acetylamino)-6-amino-2,3,6-trideoxy-alpha-D-ribo-hexopyranosyl]oxy}-4,6-diamino-2-hydroxycycloh exyl 3-amino-3-deoxy-alpha-D-glucopyranoside, ACETATE ION, Aminoglycoside 2'-N-acetyltransferase, ... | | Authors: | Bassenden, A.V, Berghuis, A.M. | | Deposit date: | 2020-02-06 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and phylogenetic analyses of resistance to next-generation aminoglycosides conferred by AAC(2') enzymes.
Sci Rep, 11, 2021
|
|
6VTA
 
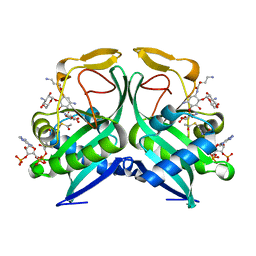 | | Aminoglycoside N-2'-Acetyltransferase-Ia [AAC(2')-Ia] in complex with amikacin and acetyl-CoA | | Descriptor: | (2S)-N-[(1R,2S,3S,4R,5S)-4-[(2R,3R,4S,5S,6R)-6-(aminomethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-5-azanyl-2-[(2S,3R,4S,5S ,6R)-4-azanyl-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]oxy-3-oxidanyl-cyclohexyl]-4-azanyl-2-oxidanyl-butanamide, ACETYL COENZYME *A, Aminoglycoside 2'-N-acetyltransferase | | Authors: | Bassenden, A.V, Berghuis, A.M. | | Deposit date: | 2020-02-12 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural and phylogenetic analyses of resistance to next-generation aminoglycosides conferred by AAC(2') enzymes.
Sci Rep, 11, 2021
|
|
7JZS
 
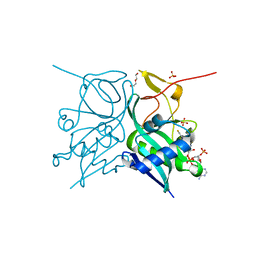 | | Aminoglycoside N-2'-Acetyltransferase-Ia [AAC(2')-Ia] in complex with CoA | | Descriptor: | Aminoglycoside 2'-N-acetyltransferase, COENZYME A, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Park, J, Bassenden, A.V, Berghuis, A.M. | | Deposit date: | 2020-09-02 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural and phylogenetic analyses of resistance to next-generation aminoglycosides conferred by AAC(2') enzymes.
Sci Rep, 11, 2021
|
|
4XJE
 
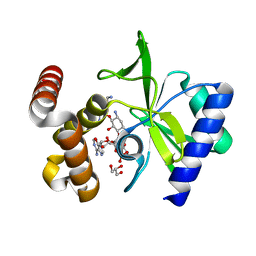 | |
5CFS
 
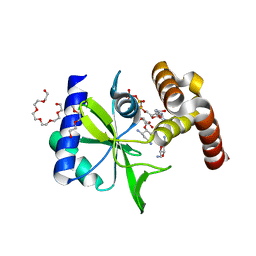 | | Crystal Structure of ANT(2")-Ia in complex with AMPCPP and tobramycin | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, AAD(2''),Gentamicin 2''-nucleotidyltransferase,Gentamicin resistance protein, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Rodionov, D, Bassenden, A.V, Berghuis, A.M. | | Deposit date: | 2015-07-08 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Analysis of the Tobramycin and Gentamicin Clinical Resistome Reveals Limitations for Next-generation Aminoglycoside Design.
Acs Chem.Biol., 11, 2016
|
|
5CFT
 
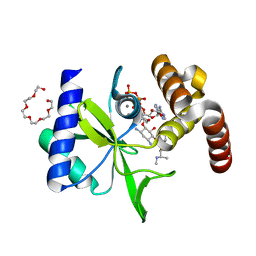 | | Crystal Structure of ANT(2")-Ia in complex with AMPCPP and gentamicin C1 | | Descriptor: | Aminoglycoside Nucleotidyltransferase (2")-Ia, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, HEXAETHYLENE GLYCOL, ... | | Authors: | Rodionov, D, Bassenden, A.V, Berghuis, A.M. | | Deposit date: | 2015-07-08 | | Release date: | 2016-03-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Analysis of the Tobramycin and Gentamicin Clinical Resistome Reveals Limitations for Next-generation Aminoglycoside Design.
Acs Chem.Biol., 11, 2016
|
|
5CFU
 
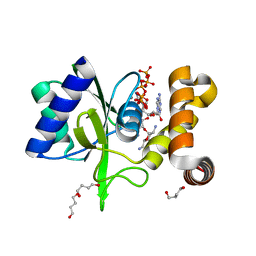 | | Crystal Structure of ANT(2")-Ia in complex with adenylyl-2"-tobramycin | | Descriptor: | 1,4-BUTANEDIOL, Aminoglycoside Nucleotidyltransferase (2")-Ia, MANGANESE (II) ION, ... | | Authors: | Rodionov, D, Bassenden, A.V, Berghuis, A.M. | | Deposit date: | 2015-07-08 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Revisiting the Catalytic Cycle and Kinetic Mechanism of AminoglycosideO-Nucleotidyltransferase(2′′): A Structural and Kinetic Study.
Acs Chem.Biol., 2020
|
|
6VOU
 
 | | Aminoglycoside N-2'-Acetyltransferase-Ia [AAC(2')-Ia] in complex with acetylated-plazomicin and CoA | | Descriptor: | (2S)-N-[(1R,2S,3S,4R,5S)-4-{[(2S,3R)-3-(acetylamino)-6-{[(2-hydroxyethyl)amino]methyl}-3,4-dihydro-2H-pyran-2-yl]oxy}-5-amino-2-{[3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranosyl]oxy}-3-hydroxycyclohexyl]-4-amino-2-hydroxybutanamide, (4S)-2-METHYL-2,4-PENTANEDIOL, 3,3',3''-phosphanetriyltripropanoic acid, ... | | Authors: | Bassenden, A.V, Berghuis, A.M. | | Deposit date: | 2020-01-31 | | Release date: | 2021-06-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for plazomicin antibiotic action and resistance.
Commun Biol, 4, 2021
|
|
5KQJ
 
 | |
7LH5
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with plazomicin, mRNA and tRNAs | | Descriptor: | (2S)-4-amino-N-[(1R,2S,3S,4R,5S)-5-amino-4-{[(2S,3R)-3-amino-6-{[(2-hydroxyethyl)amino]methyl}-3,4-dihydro-2H-pyran-2-y l]oxy}-2-{[3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranosyl]oxy}-3-hydroxycyclohexyl]-2-hydroxybutanamide, 16S ribosomal RNA, 23S ribosomal RNA, ... | | Authors: | Golkar, T, Berghuis, A.M, Schmeing, T.M. | | Deposit date: | 2021-01-21 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Structural basis for plazomicin antibiotic action and resistance.
Commun Biol, 4, 2021
|
|
